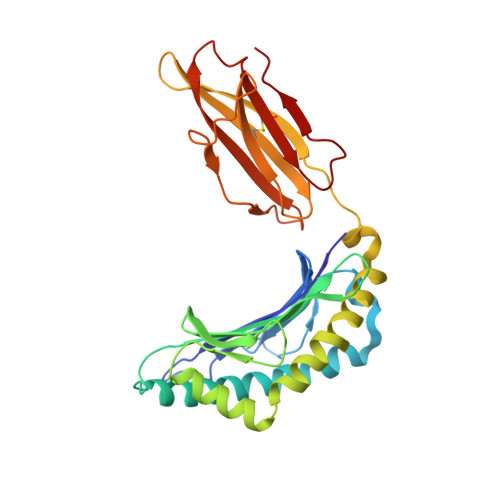
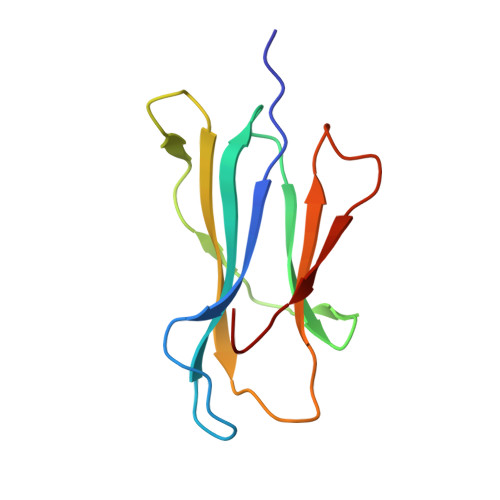
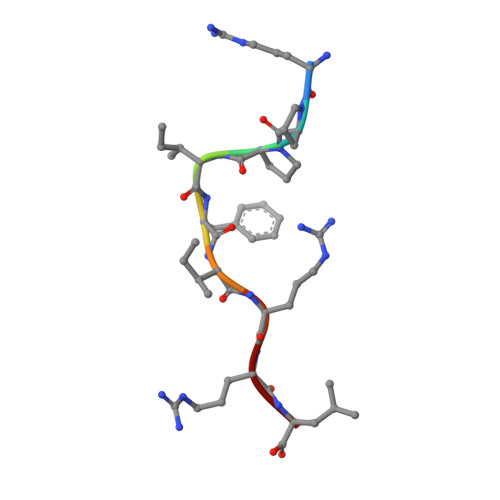
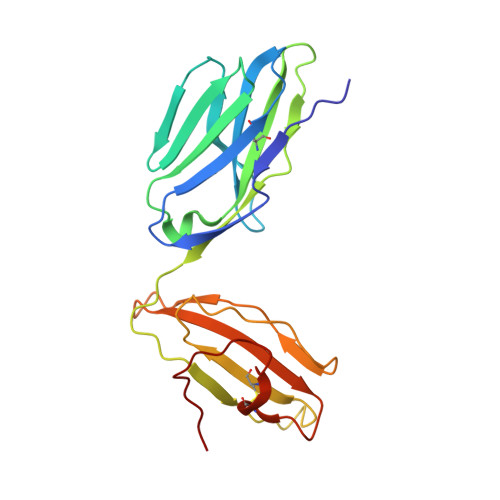
A Shared TCR Bias toward an Immunogenic EBV Epitope Dominates in HLA-B*07:02-Expressing Individuals.
Rowntree, L.C., Nguyen, T.H.O., Farenc, C., Halim, H., Hensen, L., Rossjohn, J., Kotsimbos, T.C., Purcell, A.W., Kedzierska, K., Gras, S., Mifsud, N.A.(2020) J Immunol 205: 1524-1534
- PubMed: 32817371
- DOI: https://doi.org/10.4049/jimmunol.2000249
- Primary Citation of Related Structures:
6VMX - PubMed Abstract:
EBV is one of the most common viruses found in humans and is prototypic of a persistent viral infection characterized by periods of latency. Across many HLA class I molecules, the latent-specific CD8 + T cell response is focused on epitopes derived from the EBNA-3 protein family. In the case of HLA-B*07:02 restriction, a highly frequent class I allele, the T cell response is dominated by an epitope spanning residues 379-387 of EBNA-3 (RPPIFIRRL [EBV RPP ]). However, little is known about either the TCR repertoire specific for this epitope or the molecular basis for this observed immunodominance. The EBV RPP CD8 + T cell response was common among both EBV-seropositive HLA-B*07:02 + healthy and immunocompromised individuals. Similar TCRs were identified in EBV RPP -specific CD8 + T cell repertoires across multiple HLA-B7 + individuals, indicating a shared Ag-driven bias in TCR usage. In particular, TRBV4-1 and TRAV38 usage was observed in five out of six individuals studied. In this study, we report the crystal structure of a TRBV4-1 + TCR-HLA-B*07:02/EBV RPP complex, which provides a molecular basis for the observed TRBV4-1 bias. These findings enhance our understanding of the CD8 + T cell response toward a common EBV determinant in HLA-B*07:02 + individuals.
Organizational Affiliation:
Department of Medicine, Monash University, Central Clinical School, The Alfred Hospital, Melbourne, Victoria 3004, Australia.