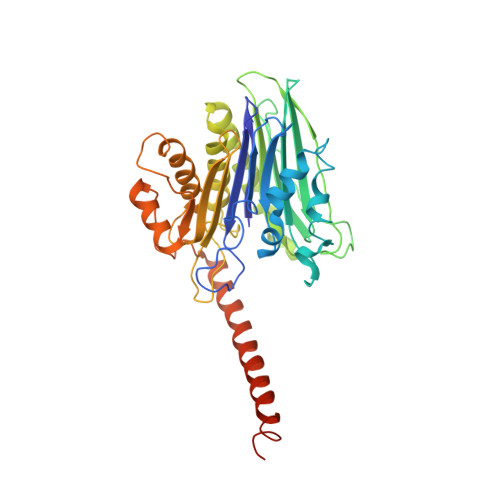
Structural insights into the function of the catalytically active human Taspase1.
Nagaratnam, N., Delker, S.L., Jernigan, R., Edwards, T.E., Snider, J., Thifault, D., Williams, D., Nannenga, B.L., Stofega, M., Sambucetti, L., Hsieh, J.J., Flint, A.J., Fromme, P., Martin-Garcia, J.M.(2021) Structure 29: 873
- PubMed: 33784495
- DOI: https://doi.org/10.1016/j.str.2021.03.008
- Primary Citation of Related Structures:
6UGK, 6VIN - PubMed Abstract:
Taspase1 is an Ntn-hydrolase overexpressed in primary human cancers, coordinating cancer cell proliferation, invasion, and metastasis. Loss of Taspase1 activity disrupts proliferation of human cancer cells in vitro and in mouse models of glioblastoma. Taspase1 is synthesized as an inactive proenzyme, becoming active upon intramolecular cleavage. The activation process changes the conformation of a long fragment at the C-terminus of the α subunit, for which no full-length structural information exists and whose function is poorly understood. We present a cloning strategy to generate a circularly permuted form of Taspase1 to determine the crystallographic structure of active Taspase1. We discovered that this region forms a long helix and is indispensable for the catalytic activity of Taspase1. Our study highlights the importance of this element for the enzymatic activity of Ntn-hydrolases, suggesting that it could be a potential target for the design of inhibitors with potential to be developed into anticancer therapeutics.
Organizational Affiliation:
Center for Applied Structural Discovery, Biodesign Institute, Arizona State University, Tempe, AZ 85287, USA.