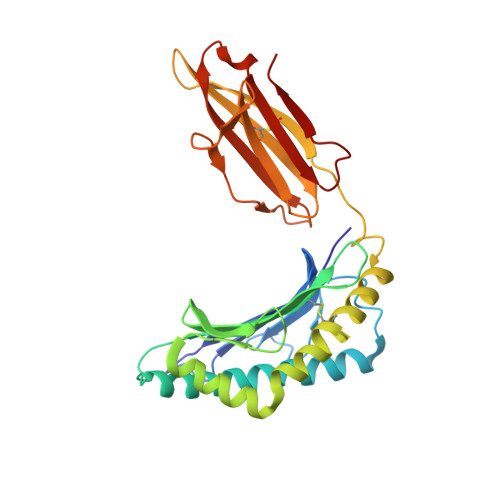
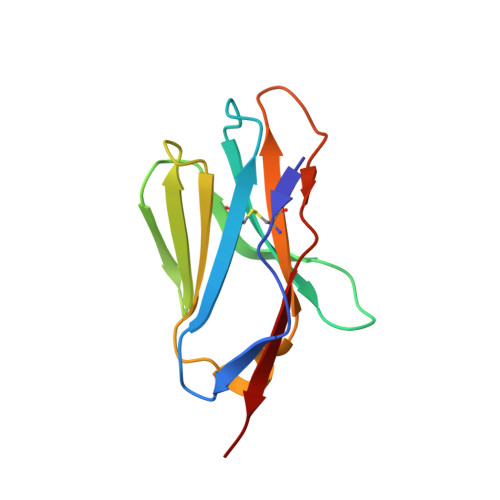
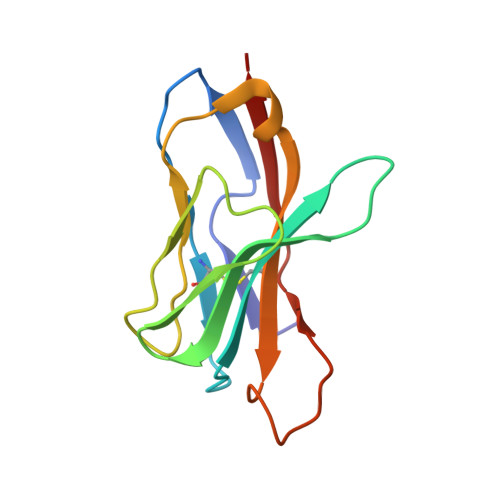
An Engineered T Cell Receptor Variant Realizes the Limits of Functional Binding Modes.
Singh, N.K., Alonso, J.A., Harris, D.T., Anderson, S.D., Ma, J., Hellman, L.M., Rosenberg, A.M., Kolawole, E.M., Evavold, B.D., Kranz, D.M., Baker, B.M.(2020) Biochemistry 59: 4163-4175
- PubMed: 33074657
- DOI: https://doi.org/10.1021/acs.biochem.0c00689
- Primary Citation of Related Structures:
6UZ1 - PubMed Abstract:
T cell receptors (TCRs) orchestrate cellular immunity by recognizing peptides presented by a range of major histocompatibility complex (MHC) proteins. Naturally occurring TCRs bind the composite peptide/MHC surface, recognizing peptides that are structurally and chemically compatible with the TCR binding site. Here we describe a molecularly evolved TCR variant that binds the human class I MHC protein HLA-A2 independent of the bound peptide, achieved by a drastic perturbation of the TCR binding geometry that places the molecule far from the peptide binding groove. This unique geometry is unsupportive of normal T cell signaling. A substantial divergence between affinity measurements in solution and in two dimensions between proximal cell membranes leads us to attribute the lack of signaling to steric hindrance that limits binding in the confines of a cell-cell interface. Our results provide an example of how receptor binding geometry can impact T cell function and provide further support for the view that germline-encoded residues in TCR binding loops evolved to drive productive TCR recognition and signaling.
Organizational Affiliation:
Department of Chemistry and Biochemistry and Harper Cancer Research Institute, University of Notre Dame, Notre Dame, Indiana 46556, United States.