An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity.
Athukoralage, J.S., McMahon, S.A., Zhang, C., Gruschow, S., Graham, S., Krupovic, M., Whitaker, R.J., Gloster, T.M., White, M.F.(2020) Nature 577: 572-575
- PubMed: 31942067
- DOI: https://doi.org/10.1038/s41586-019-1909-5
- Primary Citation of Related Structures:
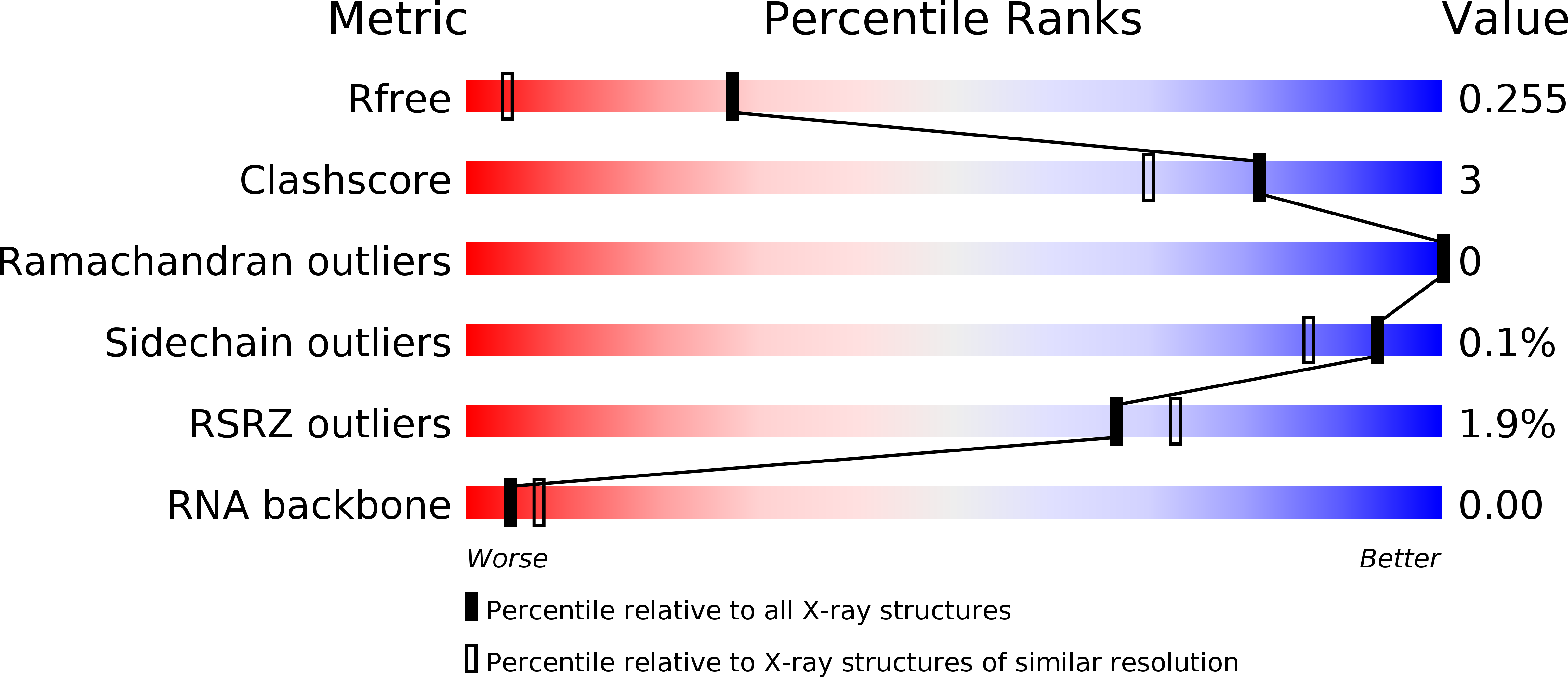
6SCF - PubMed Abstract:
The CRISPR system in bacteria and archaea provides adaptive immunity against mobile genetic elements. Type III CRISPR systems detect viral RNA, resulting in the activation of two regions of the Cas10 protein: an HD nuclease domain (which degrades viral DNA) 1,2 and a cyclase domain (which synthesizes cyclic oligoadenylates from ATP) 3-5 . Cyclic oligoadenylates in turn activate defence enzymes with a CRISPR-associated Rossmann fold domain 6 , sculpting a powerful antiviral response 7-10 that can drive viruses to extinction 7,8 . Cyclic nucleotides are increasingly implicated in host-pathogen interactions 11-13 . Here we identify a new family of viral anti-CRISPR (Acr) enzymes that rapidly degrade cyclic tetra-adenylate (cA 4 ). The viral ring nuclease AcrIII-1 is widely distributed in archaeal and bacterial viruses and in proviruses. The enzyme uses a previously unknown fold to bind cA 4 specifically, and a conserved active site to rapidly cleave this signalling molecule, allowing viruses to neutralize the type III CRISPR defence system. The AcrIII-1 family has a broad host range, as it targets cA 4 signalling molecules rather than specific CRISPR effector proteins. Our findings highlight the crucial role of cyclic nucleotide signalling in the conflict between viruses and their hosts.
Organizational Affiliation:
Biomedical Sciences Research Complex, School of Biology, University of St Andrews, St Andrews, Fife, UK.