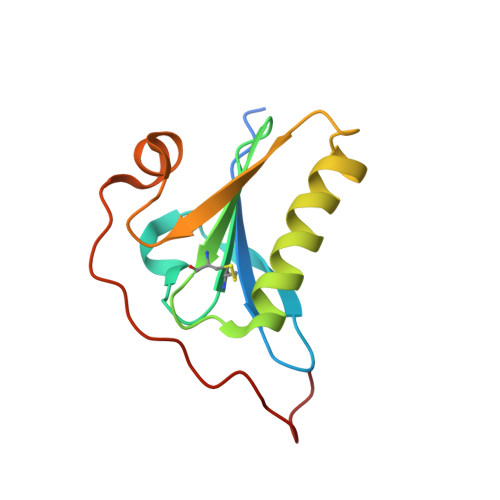
High-resolution crystal structure of gelsolin domain 2 in complex with the physiological calcium ion.
Bollati, M., Scalone, E., Boni, F., Mastrangelo, E., Giorgino, T., Milani, M., de Rosa, M.(2019) Biochem Biophys Res Commun 518: 94-99
- PubMed: 31416615
- DOI: https://doi.org/10.1016/j.bbrc.2019.08.013
- Primary Citation of Related Structures:
6QW3 - PubMed Abstract:
The second domain of gelsolin (G2) hosts mutations responsible for a hereditary form of amyloidosis. The active form of gelsolin is Ca 2+ -bound; it is also a dynamic protein, hence structural biologists often rely on the study of the isolated G2. However, the wild type G2 structure that have been used so far in comparative studies is bound to a crystallographic Cd 2+ , in lieu of the physiological calcium. Here, we report the wild type structure of G2 in complex with Ca 2+ highlighting subtle ion-dependent differences. Previous findings on different G2 mutations are also briefly revised in light of these results.
Organizational Affiliation:
Istituto di Biofisica, Consiglio Nazionale delle Ricerche, Via Celoria 26, 20133, Milano, Italy; Dipartimento di Bioscienze, Università degli Studi di Milano, Via Celoria 26, 20133, Milano, Italy.