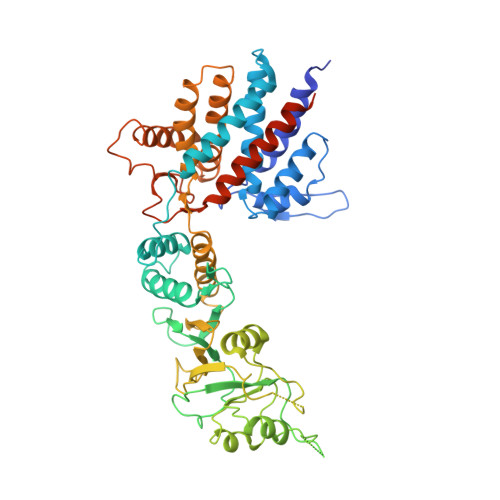
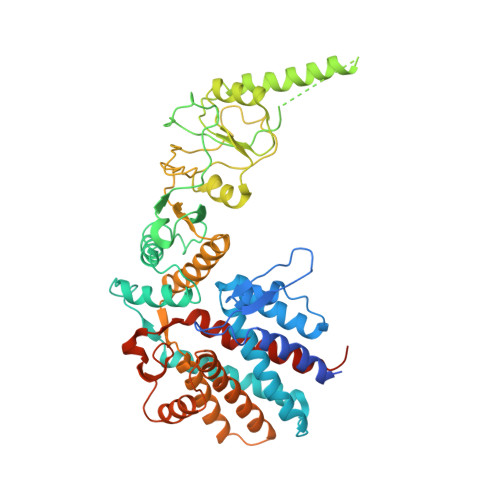
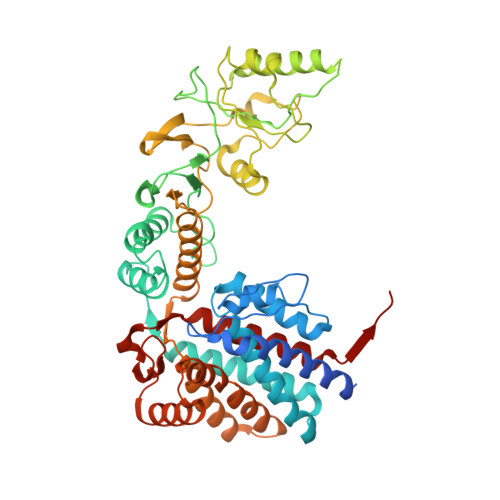
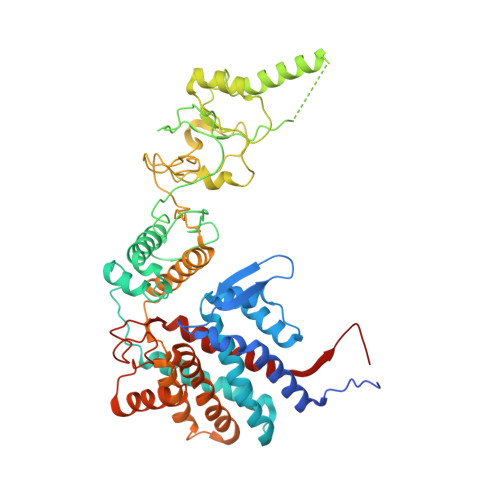
Structural and functional analysis of the role of the chaperonin CCT in mTOR complex assembly.
Cuellar, J., Ludlam, W.G., Tensmeyer, N.C., Aoba, T., Dhavale, M., Santiago, C., Bueno-Carrasco, M.T., Mann, M.J., Plimpton, R.L., Makaju, A., Franklin, S., Willardson, B.M., Valpuesta, J.M.(2019) Nat Commun 10: 2865-2865
- PubMed: 31253771
- DOI: https://doi.org/10.1038/s41467-019-10781-1
- Primary Citation of Related Structures:
6QB8 - PubMed Abstract:
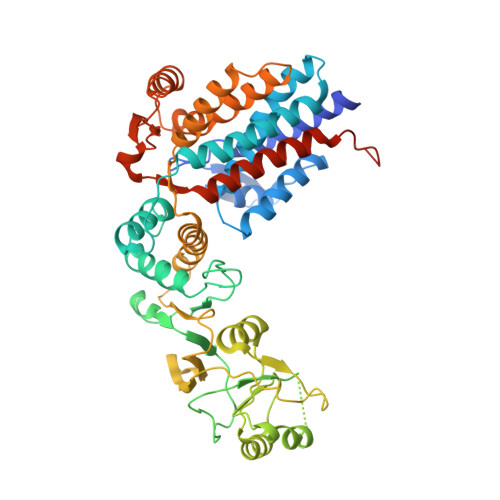
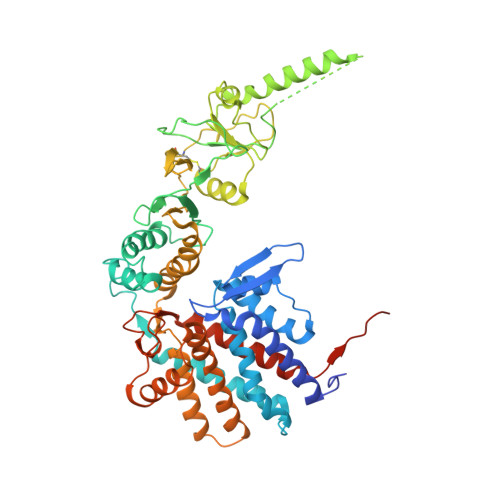
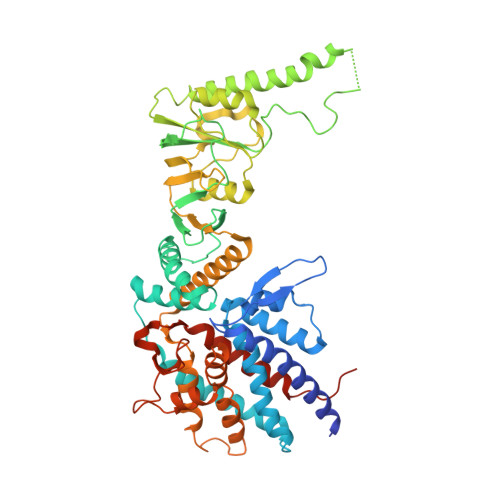
The mechanistic target of rapamycin (mTOR) kinase forms two multi-protein signaling complexes, mTORC1 and mTORC2, which are master regulators of cell growth, metabolism, survival and autophagy. Two of the subunits of these complexes are mLST8 and Raptor, β-propeller proteins that stabilize the mTOR kinase and recruit substrates, respectively. Here we report that the eukaryotic chaperonin CCT plays a key role in mTORC assembly and signaling by folding both mLST8 and Raptor. A high resolution (4.0 Å) cryo-EM structure of the human mLST8-CCT intermediate isolated directly from cells shows mLST8 in a near-native state bound to CCT deep within the folding chamber between the two CCT rings, and interacting mainly with the disordered N- and C-termini of specific CCT subunits of both rings. These findings describe a unique function of CCT in mTORC assembly and a distinct binding site in CCT for mLST8, far from those found for similar β-propeller proteins.
Organizational Affiliation:
Centro Nacional de Biotecnología, Campus de la Universidad Autónoma de Madrid, 28049, Madrid, Spain.