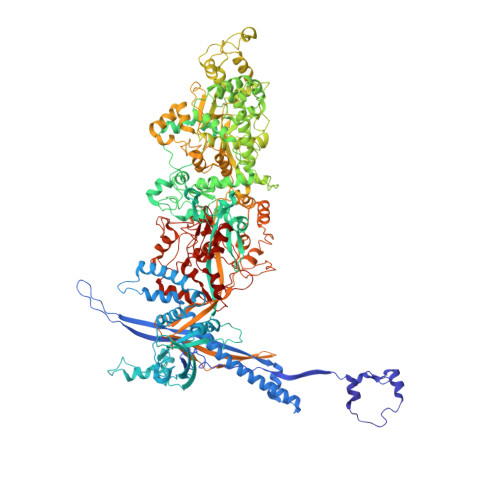
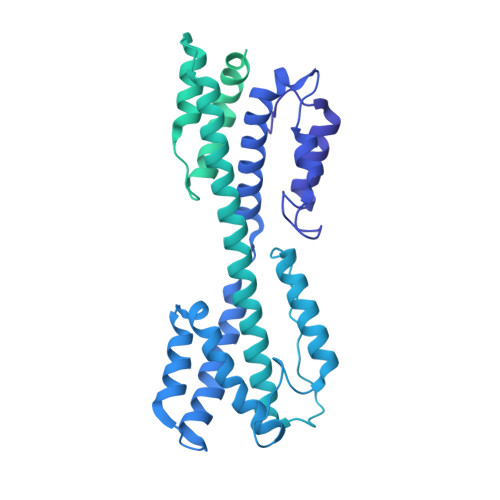
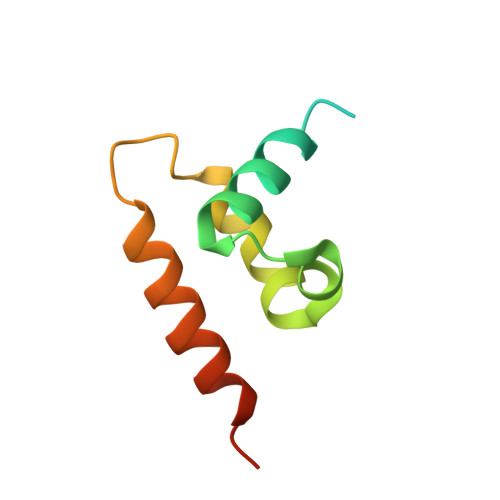
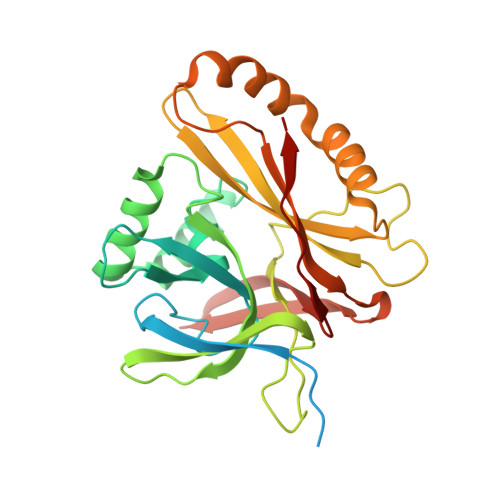
Atomic structure of the human herpesvirus 6B capsid and capsid-associated tegument complexes.
Zhang, Y., Liu, W., Li, Z., Kumar, V., Alvarez-Cabrera, A.L., Leibovitch, E.C., Cui, Y., Mei, Y., Bi, G.Q., Jacobson, S., Zhou, Z.H.(2019) Nat Commun 10: 5346-5346
- PubMed: 31767868
- DOI: https://doi.org/10.1038/s41467-019-13064-x
- Primary Citation of Related Structures:
6Q1F - PubMed Abstract:
Human herpesvirus 6B (HHV-6B) belongs to the β-herpesvirus subfamily of the Herpesviridae. To understand capsid assembly and capsid-tegument interactions, here we report atomic structures of HHV-6B capsid and capsid-associated tegument complex (CATC) obtained by cryoEM and sub-particle reconstruction. Compared to other β-herpesviruses, HHV-6B exhibits high similarity in capsid structure but organizational differences in its CATC (pU11 tetramer). 180 "VΛ"-shaped CATCs are observed in HHV-6B, distinguishing from the 255 "Λ"-shaped dimeric CATCs observed in murine cytomegalovirus and the 310 "Δ"-shaped CATCs in human cytomegalovirus. This trend in CATC quantity correlates with the increasing genomes sizes of these β-herpesviruses. Incompatible distances revealed by the atomic structures rationalize the lack of CATC's binding to triplexes Ta, Tc, and Tf in HHV-6B. Our results offer insights into HHV-6B capsid assembly and the roles of its tegument proteins, including not only the β-herpesvirus-specific pU11 and pU14, but also those conserved across all subfamilies of Herpesviridae.
Organizational Affiliation:
Center for Integrative Imaging, Hefei National Laboratory for Physical Sciences at the Microscale, and School of Life Sciences, University of Science and Technology of China (USTC), Hefei, Anhui, 230026, China.