6PV8
Human alpha3beta4 nicotinic acetylcholine receptor in complex with AT-1001
- PDB DOI: https://doi.org/10.2210/pdb6PV8/pdb
- EM Map EMD-20488: EMDB EMDataResource
- Classification: MEMBRANE PROTEIN
- Organism(s): Homo sapiens, Escherichia coli O11, Mus musculus
- Expression System: Homo sapiens
- Mutation(s): No
- Membrane Protein: Yes PDBTMMemProtMDmpstruc
- Deposited: 2019-07-19 Released: 2019-09-11
- Funding Organization(s): Welch Foundation, National Institutes of Health/National Institute on Drug Abuse (NIH/NIDA), National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS), National Institutes of Health/National Institute of Neurological Disorders and Stroke (NIH/NINDS)
Experimental Data Snapshot
- Method: ELECTRON MICROSCOPY
- Resolution: 3.87 Å
- Aggregation State: PARTICLE
- Reconstruction Method: SINGLE PARTICLE
wwPDB Validation 3D Report Full Report
This is version 2.0 of the entry. See complete history.
Macromolecules
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fusion protein of Neuronal acetylcholine receptor subunit alpha-3 and Soluble cytochrome b562 | 525 | Homo sapiens, Escherichia coli O11 | Mutation(s): 0 Gene Names: CHRNA3, NACHRA3, CWJ32_12820 Membrane Entity: Yes |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P32297 (Homo sapiens) Explore P32297 Go to UniProtKB: P32297 | |||||
PHAROS: P32297 GTEx: ENSG00000080644 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P32297 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fusion protein of Neuronal acetylcholine receptor subunit beta-4 and Soluble cytochrome b562 | 538 | Homo sapiens, Escherichia coli O11 | Mutation(s): 0 Gene Names: CHRNB4, CWJ32_12820 Membrane Entity: Yes | 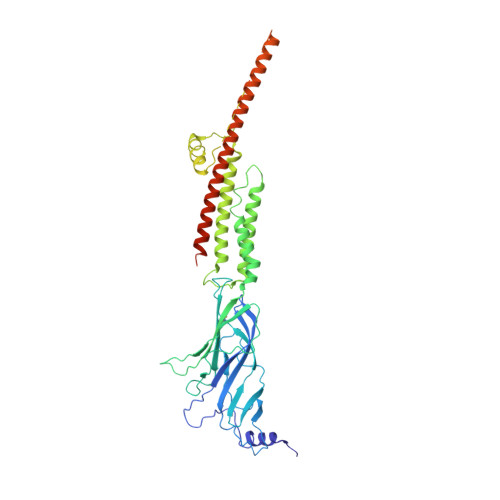 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P30926 (Homo sapiens) Explore P30926 Go to UniProtKB: P30926 | |||||
PHAROS: P30926 GTEx: ENSG00000117971 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P30926 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| IgG2b Fab heavy chain | 219 | Mus musculus | Mutation(s): 0 | 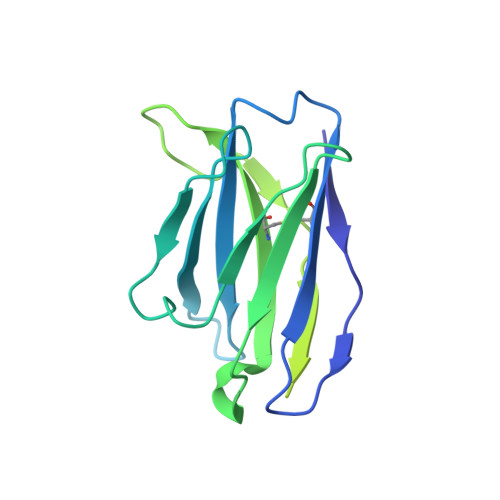 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Kappa Fab light chain | 213 | Mus musculus | Mutation(s): 0 | 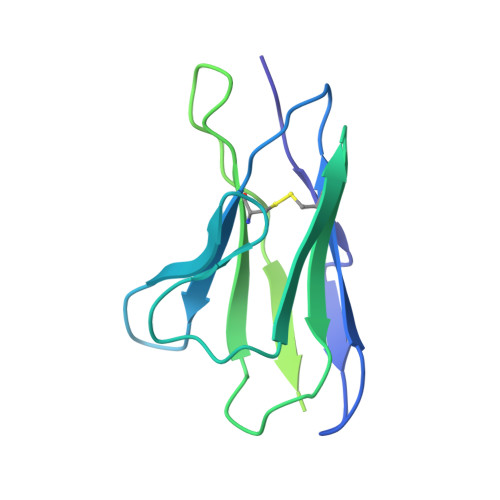 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Oligosaccharides
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | J, P | 2 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G42666HT GlyCosmos: G42666HT GlyGen: G42666HT | |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | K, M, O, Q, S | 5 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G22768VO GlyCosmos: G22768VO GlyGen: G22768VO | |||||
Small Molecules
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| Y01 Query on Y01 | DA [auth C] EA [auth C] GA [auth D] HA [auth D] KA [auth E] | CHOLESTEROL HEMISUCCINATE C31 H50 O4 WLNARFZDISHUGS-MIXBDBMTSA-N |  | ||
| P1M (Subject of Investigation/LOI) Query on P1M | FA [auth D], T [auth A] | (3-endo)-N-(2-bromophenyl)-9-methyl-9-azabicyclo[3.3.1]nonan-3-amine C15 H21 Br N2 UZJWAFOJOSGEKL-CLLJXQQHSA-N |  | ||
| NAG Query on NAG | BA [auth C] CA [auth C] IA [auth E] JA [auth E] W [auth B] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| NA Query on NA | AA [auth B] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
Experimental Data & Validation
Experimental Data
- Method: ELECTRON MICROSCOPY
- Resolution: 3.87 Å
- Aggregation State: PARTICLE
- Reconstruction Method: SINGLE PARTICLE
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | RELION | |
| MODEL REFINEMENT | PHENIX |
Entry History & Funding Information
Deposition Data
- Released Date: 2019-09-11 Deposition Author(s): Gharpure, A., Teng, J., Zhuang, Y., Noviello, C.M., Walsh, R.M., Cabuco, R., Howard, R.J., Zaveri, N.T., Lindahl, E., Hibbs, R.E.
| Funding Organization | Location | Grant Number |
|---|---|---|
| Welch Foundation | United States | I-1812 |
| National Institutes of Health/National Institute on Drug Abuse (NIH/NIDA) | United States | T32DA07290 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | T32GM008203 |
| National Institutes of Health/National Institute on Drug Abuse (NIH/NIDA) | United States | DA037492 |
| National Institutes of Health/National Institute on Drug Abuse (NIH/NIDA) | United States | DA042072 |
| National Institutes of Health/National Institute of Neurological Disorders and Stroke (NIH/NINDS) | United States | NS095899 |
Revision History (Full details and data files)
- Version 1.0: 2019-09-11
Type: Initial release - Version 1.1: 2019-09-18
Changes: Data collection, Database references - Version 1.2: 2019-11-20
Changes: Database references - Version 1.3: 2019-12-11
Changes: Author supporting evidence - Version 2.0: 2020-07-29
Type: Remediation
Reason: Carbohydrate remediation
Changes: Advisory, Atomic model, Data collection, Derived calculations, Structure summary














