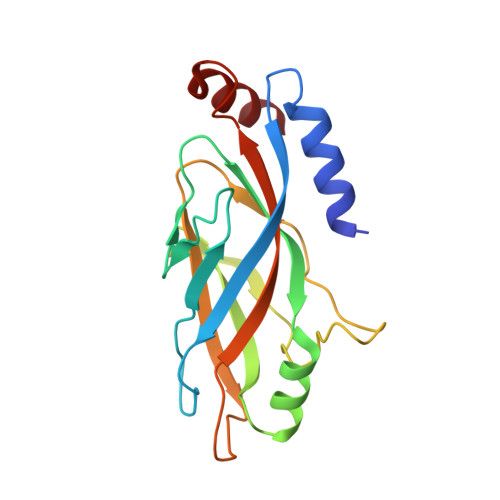
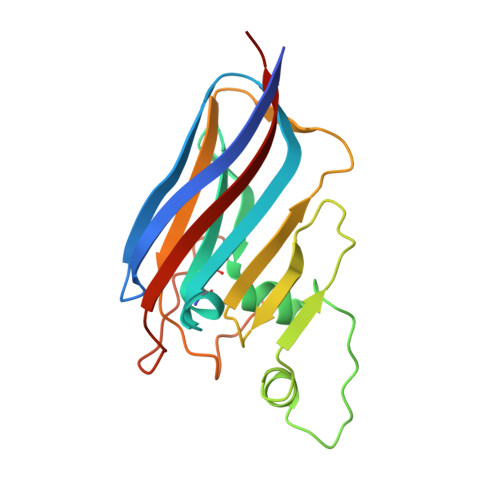
A packing for A-form DNA in an icosahedral virus.
Wang, F., Liu, Y., Su, Z., Osinski, T., de Oliveira, G.A.P., Conway, J.F., Schouten, S., Krupovic, M., Prangishvili, D., Egelman, E.H.(2019) Proc Natl Acad Sci U S A 116: 22591-22597
- PubMed: 31636205
- DOI: https://doi.org/10.1073/pnas.1908242116
- Primary Citation of Related Structures:
6OJ0 - PubMed Abstract:
Studies on viruses infecting archaea living in the most extreme environments continue to show a remarkable diversity of structures, suggesting that the sampling continues to be very sparse. We have used electron cryo-microscopy to study at 3.7-Å resolution the structure of the Sulfolobus polyhedral virus 1 (SPV1), which was originally isolated from a hot, acidic spring in Beppu, Japan. The 2 capsid proteins with variant single jelly-roll folds form pentamers and hexamers which assemble into a T = 43 icosahedral shell. In contrast to tailed icosahedral double-stranded DNA (dsDNA) viruses infecting bacteria and archaea, and herpesviruses infecting animals and humans, where naked DNA is packed under very high pressure due to the repulsion between adjacent layers of DNA, the circular dsDNA in SPV1 is fully covered with a viral protein forming a nucleoprotein filament with attractive interactions between layers. Most strikingly, we have been able to show that the DNA is in an A-form, as it is in the filamentous viruses infecting hyperthermophilic acidophiles. Previous studies have suggested that DNA is in the B-form in bacteriophages, and our study is a direct visualization of the structure of DNA in an icosahedral virus.
Organizational Affiliation:
Department of Biochemistry and Molecular Genetics, University of Virginia, Charlottesville, VA 22908.