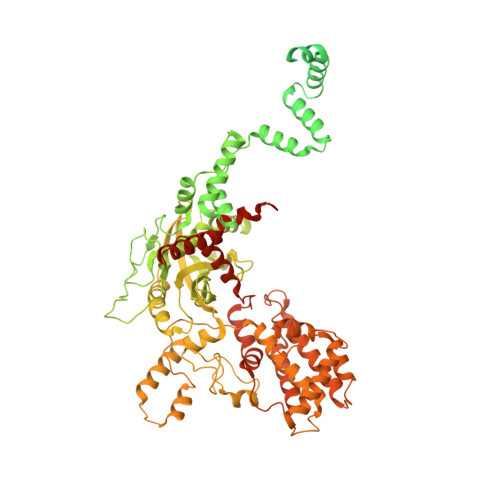
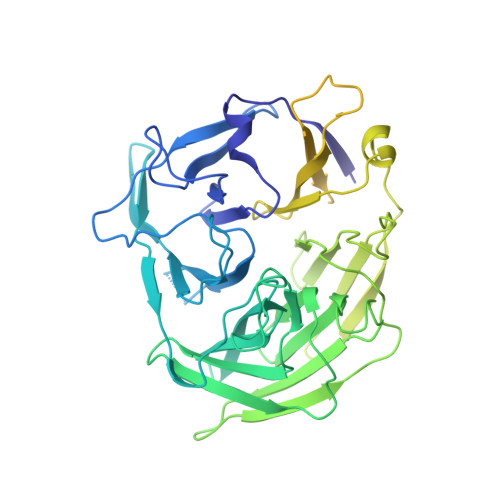
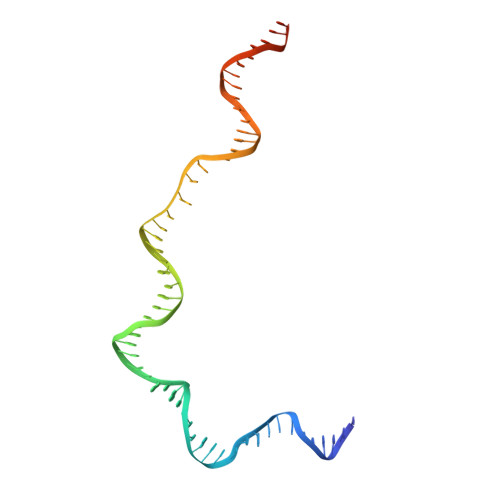
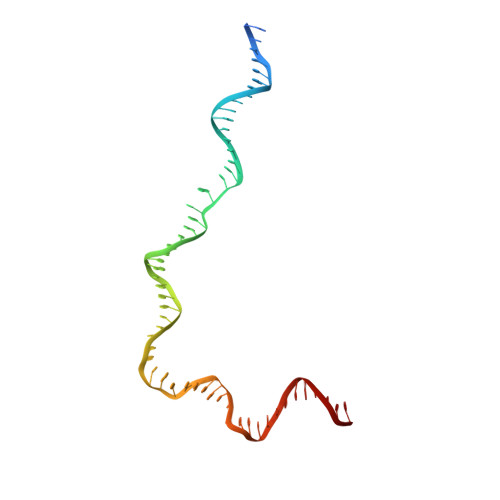
Cutting antiparallel DNA strands in a single active site.
Chen, X., Cui, Y., Best, R.B., Wang, H., Zhou, Z.H., Yang, W., Gellert, M.(2020) Nat Struct Mol Biol 27: 119-126
- PubMed: 32015552
- DOI: https://doi.org/10.1038/s41594-019-0363-2
- Primary Citation of Related Structures:
6OEM, 6OEN, 6OEO, 6OEP, 6OEQ, 6OER, 6V0V - PubMed Abstract:
A single enzyme active site that catalyzes multiple reactions is a well-established biochemical theme, but how one nuclease site cleaves both DNA strands of a double helix has not been well understood. In analyzing site-specific DNA cleavage by the mammalian RAG1-RAG2 recombinase, which initiates V(D)J recombination, we find that the active site is reconfigured for the two consecutive reactions and the DNA double helix adopts drastically different structures. For initial nicking of the DNA, a locally unwound and unpaired DNA duplex forms a zipper via alternating interstrand base stacking, rather than melting as generally thought. The second strand cleavage and formation of a hairpin-DNA product requires a global scissor-like movement of protein and DNA, delivering the scissile phosphate into the rearranged active site.
Organizational Affiliation:
Laboratory of Molecular Biology, NIDDK, National Institutes of Health, Bethesda, MD, USA.