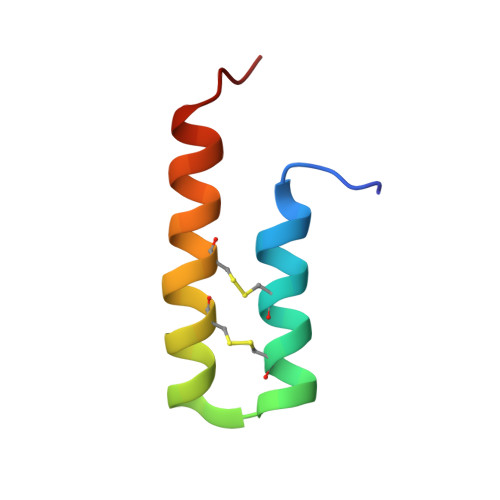
An Ancient Peptide Family Buried within Vicilin Precursors.
Zhang, J., Payne, C.D., Pouvreau, B., Schaefer, H., Fisher, M.F., Taylor, N.L., Berkowitz, O., Whelan, J., Rosengren, K.J., Mylne, J.S.(2019) ACS Chem Biol 14: 979-993
- PubMed: 30973714
- DOI: https://doi.org/10.1021/acschembio.9b00167
- Primary Citation of Related Structures:
6O3Q, 6O3S - PubMed Abstract:
New proteins can evolve by duplication and divergence or de novo, from previously noncoding DNA. A recently observed mechanism is for peptides to evolve within a "host" protein and emerge by proteolytic processing. The first examples of such interstitial peptides were ones hosted by precursors for seed storage albumin. Interstitial peptides have also been observed in precursors for seed vicilins, but current evidence for vicilin-buried peptides (VBPs) is limited to seeds of the broadleaf plants pumpkin and macadamia. Here, an extensive sequence analysis of vicilin precursors suggested that peptides buried within the N-terminal region of preprovicilins are widespread and truly ancient. Gene sequences indicative of interstitial peptides were found in species from Amborellales to eudicots and include important grass and legume crop species. We show the first protein evidence for a monocot VBP in date palm seeds as well as protein evidence from other crops including the common tomato, sesame and pumpkin relatives, cucumber, and the sponge loofah ( Luffa aegyptiaca). Their excision was consistent with asparaginyl endopeptidase-mediated maturation, and sequences were confirmed by tandem mass spectrometry. Our findings suggest that the family is large and ancient and that based on the NMR solution structures for loofah Luffin P1 and tomato VBP-8, VBPs adopt a helical hairpin fold stapled by two internal disulfide bonds. The first VBPs characterized were a protease inhibitor, antimicrobials, and a ribosome inactivator. The age and evolutionary retention of this peptide family suggest its members play important roles in plant biology.
Organizational Affiliation:
Faculty of Medicine, School of Biomedical Sciences , The University of Queensland , Brisbane , Queensland 4072 , Australia.