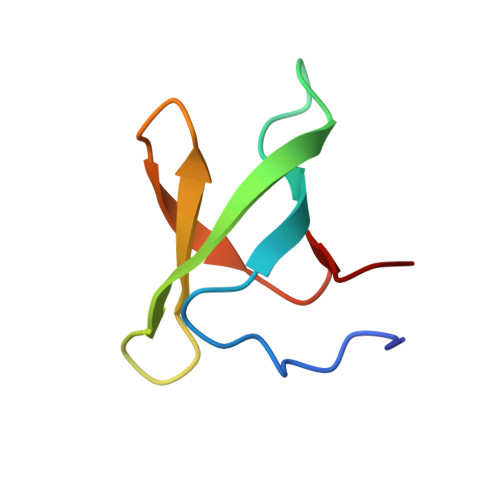
The structure of a highly-conserved picocyanobacterial protein reveals a Tudor domain with an RNA-binding function.
Bauer, K.M., Dicovitsky, R., Pellegrini, M., Zhaxybayeva, O., Ragusa, M.J.(2019) J Biol Chem 294: 14333-14344
- PubMed: 31391250
- DOI: https://doi.org/10.1074/jbc.RA119.007938
- Primary Citation of Related Structures:
6NNB - PubMed Abstract:
Cyanobacteria of the Prochlorococcus and marine Synechococcus genera are the most abundant photosynthetic microbes in the ocean. Intriguingly, the genomes of these bacteria are strongly divergent even within each genus, both in gene content and at the amino acid level of the encoded proteins. One striking exception to this is a 62-amino-acid protein, termed P rochlorococcus/ S ynechococcus h yper- c onserved p rotein (PSHCP). PSHCP is not only found in all sequenced Prochlorococcus and marine Synechococcus genomes, but it is also nearly 100% identical in its amino acid sequence across all sampled genomes. Such universal distribution and sequence conservation suggest an essential cellular role of PSHCP in these bacteria. However, its function is unknown. Here, we used NMR spectroscopy to determine its structure, finding that 53 of the 62 amino acids in PSHCP form a Tudor domain, whereas the remainder of the protein is disordered. NMR titration experiments revealed that PSHCP has only a weak affinity for DNA, but an 18.5-fold higher affinity for tRNA, hinting at an involvement of PSHCP in translation. Isothermal titration calorimetry experiments further revealed that PSHCP also binds single-stranded, double-stranded, and hairpin RNAs. These results provide the first insight into the structure and function of PSHCP, suggesting that PSHCP appears to be an RNA-binding protein that can recognize a broad array of RNA molecules.
Organizational Affiliation:
Department of Biochemistry and Cell Biology, Geisel School of Medicine, Dartmouth College, Hanover, New Hampshire 03755.