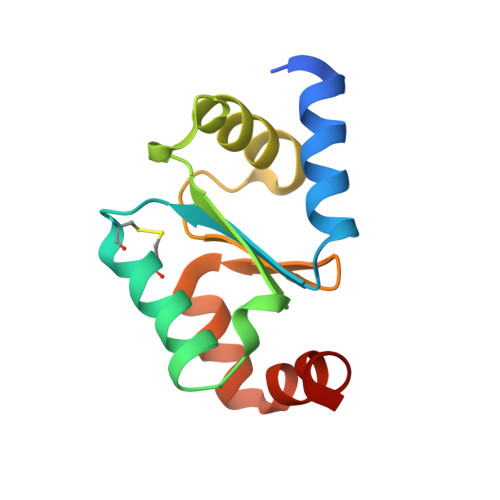
High-resolution crystal structure of the reduced Grx1 from Saccharomyces cerevisiae.
Maghool, S., La Fontaine, S., Maher, M.J.(2019) Acta Crystallogr F Struct Biol Commun 75: 392-396
- PubMed: 31045569
- DOI: https://doi.org/10.1107/S2053230X19003327
- Primary Citation of Related Structures:
6MWS - PubMed Abstract:
Grx1, a cytosolic thiol-disulfide oxidoreductase, actively maintains cellular redox homeostasis using glutathione substrates (reduced, GSH, and oxidized, GSSG). Here, the crystallization of reduced Grx1 from the yeast Saccharomyces cerevisiae (yGrx1) in space group P2 1 2 1 2 1 and its structure solution and refinement to 1.22 Å resolution are reported. To study the structure-function relationship of yeast Grx1, the crystal structure of reduced yGrx1 was compared with the existing structures of the oxidized and glutathionylated forms. These comparisons revealed structural differences in the conformations of residues neighbouring the Cys27-Cys30 active site which accompany alterations in the redox status of the protein.
Organizational Affiliation:
Department of Biochemistry and Genetics, La Trobe Institute for Molecular Science, La Trobe University, Melbourne, Victoria, Australia.