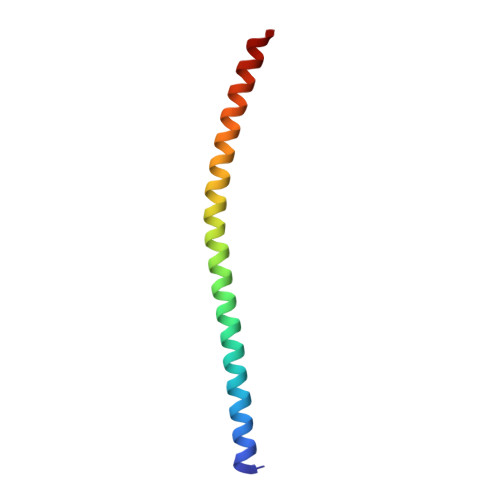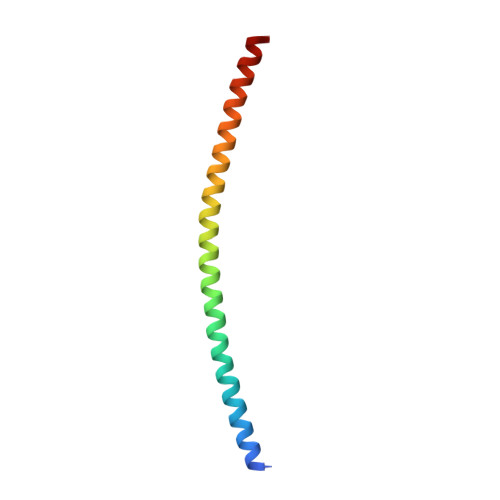Structural basis for the clamping and Ca2+activation of SNARE-mediated fusion by synaptotagmin.
Grushin, K., Wang, J., Coleman, J., Rothman, J.E., Sindelar, C.V., Krishnakumar, S.S.(2019) Nat Commun 10: 2413-2413
- PubMed: 31160571
- DOI: https://doi.org/10.1038/s41467-019-10391-x
- Primary Citation of Related Structures:
6MTI - PubMed Abstract:
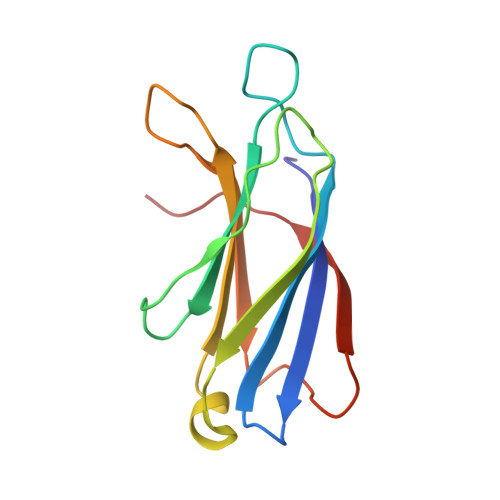
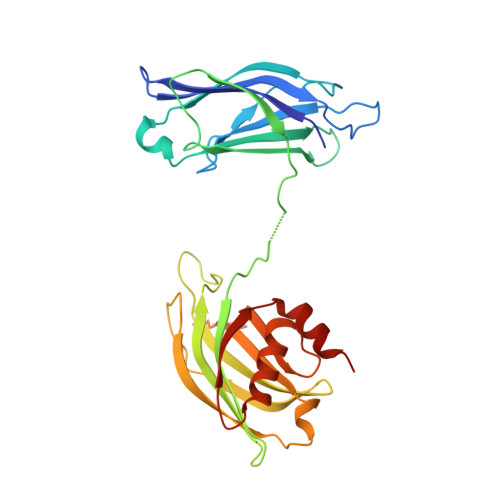
Synapotagmin-1 (Syt1) interacts with both SNARE proteins and lipid membranes to synchronize neurotransmitter release to calcium (Ca 2+ ) influx. Here we report the cryo-electron microscopy structure of the Syt1-SNARE complex on anionic-lipid containing membranes. Under resting conditions, the Syt1 C2 domains bind the membrane with a magnesium (Mg 2+ )-mediated partial insertion of the aliphatic loops, alongside weak interactions with the anionic lipid headgroups. The C2B domain concurrently interacts the SNARE bundle via the 'primary' interface and is positioned between the SNAREpins and the membrane. In this configuration, Syt1 is projected to sterically delay the complete assembly of the associated SNAREpins and thus, contribute to clamping fusion. This Syt1-SNARE organization is disrupted upon Ca 2+ -influx as Syt1 reorients into the membrane, likely displacing the attached SNAREpins and reversing the fusion clamp. We thus conclude that the cation (Mg 2+ /Ca 2+ ) dependent membrane interaction is a key determinant of the dual clamp/activator function of Synaptotagmin-1.
Organizational Affiliation:
Department of Cell Biology, Yale University School of Medicine, 333 Cedar Street, New Haven, CT, 06520, USA.