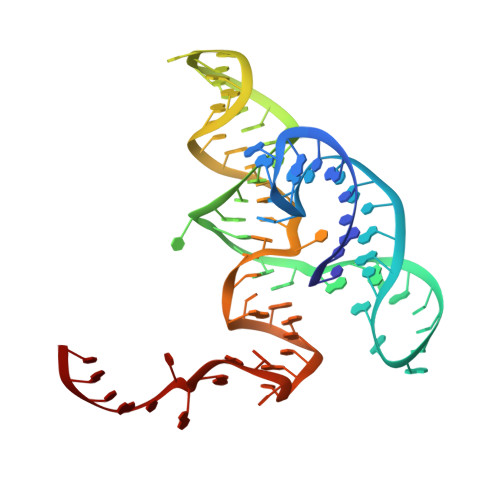
Hatchet ribozyme structure and implications for cleavage mechanism.
Zheng, L., Falschlunger, C., Huang, K., Mairhofer, E., Yuan, S., Wang, J., Patel, D.J., Micura, R., Ren, A.(2019) Proc Natl Acad Sci U S A 116: 10783-10791
- PubMed: 31088965
- DOI: https://doi.org/10.1073/pnas.1902413116
- Primary Citation of Related Structures:
6JQ5, 6JQ6 - PubMed Abstract:
Small self-cleaving ribozymes catalyze site-specific cleavage of their own phosphodiester backbone with implications for viral genome replication, pre-mRNA processing, and alternative splicing. We report on the 2.1-Å crystal structure of the hatchet ribozyme product, which adopts a compact pseudosymmetric dimeric scaffold, with each monomer stabilized by long-range interactions involving highly conserved nucleotides brought into close proximity of the scissile phosphate. Strikingly, the catalytic pocket contains a cavity capable of accommodating both the modeled scissile phosphate and its flanking 5' nucleoside. The resulting modeled precatalytic conformation incorporates a splayed-apart alignment at the scissile phosphate, thereby providing structure-based insights into the in-line cleavage mechanism. We identify a guanine lining the catalytic pocket positioned to contribute to cleavage chemistry. The functional relevance of structure-based insights into hatchet ribozyme catalysis is strongly supported by cleavage assays monitoring the impact of selected nucleobase and atom-specific mutations on ribozyme activity.
Organizational Affiliation:
Life Science Institute, Zhejiang University, 310058 Hangzhou, China.