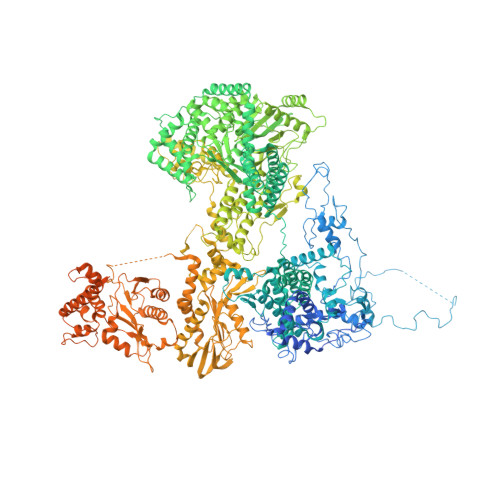
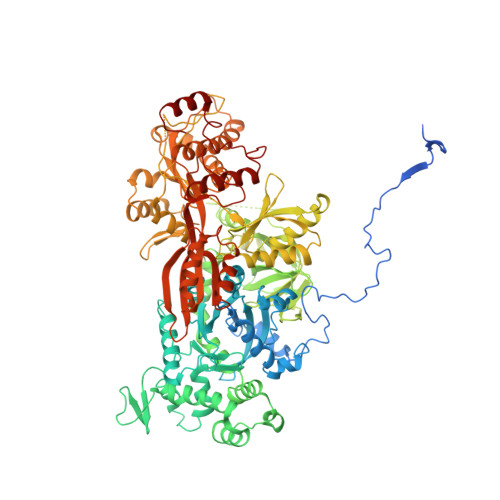
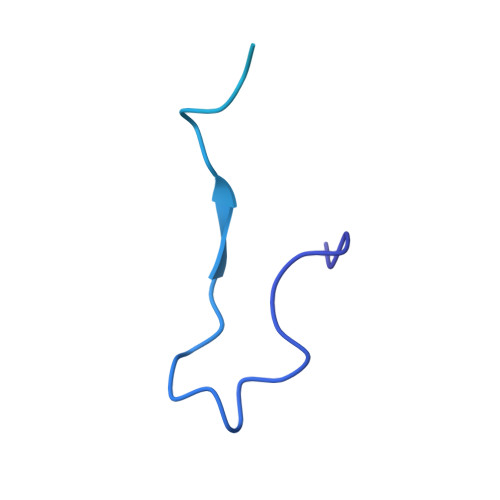
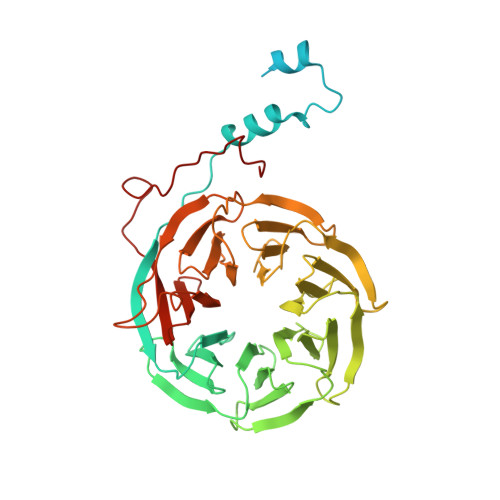
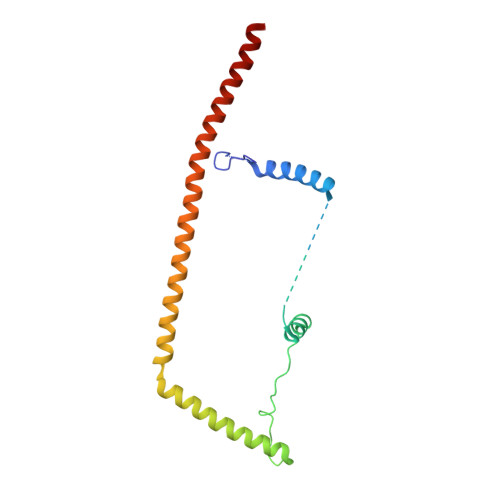
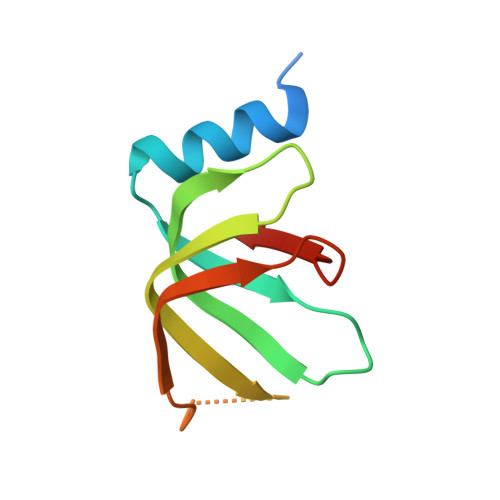
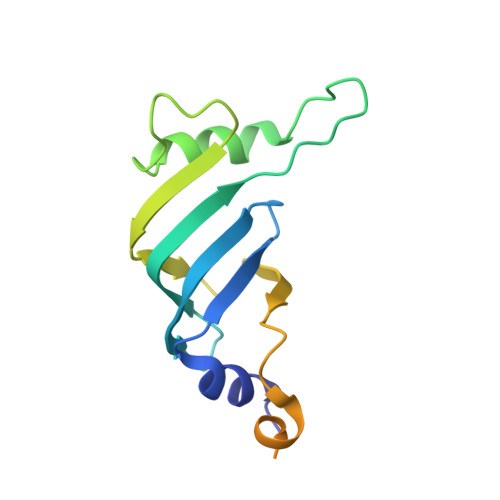
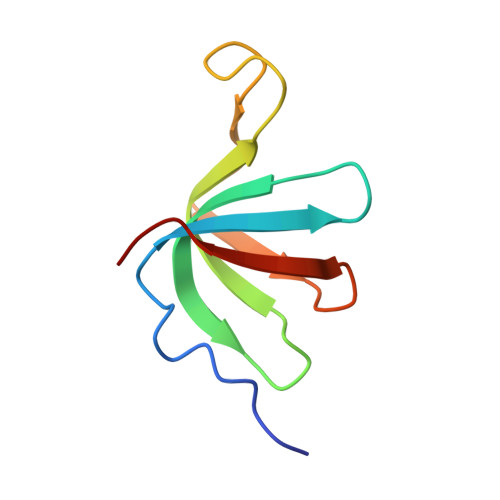
Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Wan, R., Bai, R., Yan, C., Lei, J., Shi, Y.(2019) Cell 177: 339-351.e13
- PubMed: 30879786
- DOI: https://doi.org/10.1016/j.cell.2019.02.006
- Primary Citation of Related Structures:
6J6G, 6J6H, 6J6N, 6J6Q - PubMed Abstract:
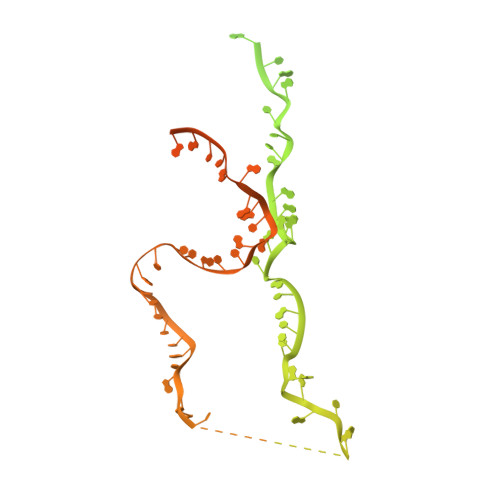
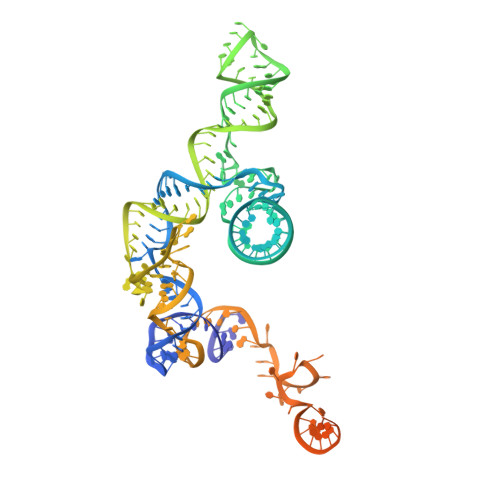
Pre-mRNA splicing is executed by the spliceosome. Structural characterization of the catalytically activated complex (B ∗ ) is pivotal for understanding the branching reaction. In this study, we assembled the B ∗ complexes on two different pre-mRNAs from Saccharomyces cerevisiae and determined the cryo-EM structures of four distinct B ∗ complexes at overall resolutions of 2.9-3.8 Å. The duplex between U2 small nuclear RNA (snRNA) and the branch point sequence (BPS) is discretely away from the 5'-splice site (5'SS) in the three B ∗ complexes that are devoid of the step I splicing factors Yju2 and Cwc25. Recruitment of Yju2 into the active site brings the U2/BPS duplex into the vicinity of 5'SS, with the BPS nucleophile positioned 4 Å away from the catalytic metal M2. This analysis reveals the functional mechanism of Yju2 and Cwc25 in branching. These structures on different pre-mRNAs reveal substrate-specific conformations of the spliceosome in a major functional state.
Organizational Affiliation:
Beijing Advanced Innovation Center for Structural Biology, Tsinghua-Peking Joint Center for Life Sciences, School of Life Sciences and School of Medicine, Tsinghua University, Beijing 100084, China.