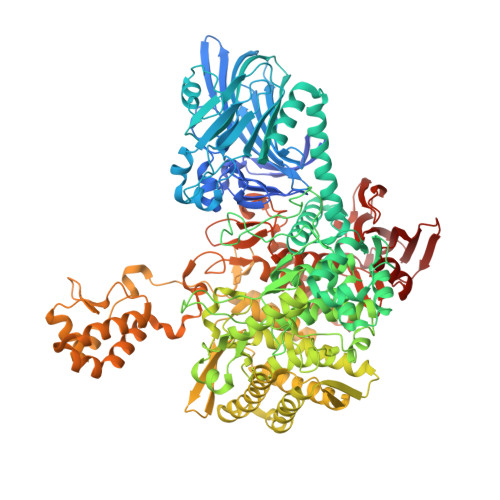
The structure of a GH149 beta-(1 → 3) glucan phosphorylase reveals a new surface oligosaccharide binding site and additional domains that are absent in the disaccharide-specific GH94 glucose-beta-(1 → 3)-glucose (laminaribiose) phosphorylase.
Kuhaudomlarp, S., Stevenson, C.E.M., Lawson, D.M., Field, R.A.(2019) Proteins 87: 885-892
- PubMed: 31134667
- DOI: https://doi.org/10.1002/prot.25745
- Primary Citation of Related Structures:
6HQ6, 6HQ8 - PubMed Abstract:
Glycoside phosphorylases (GPs) with specificity for β-(1 → 3)-gluco-oligosaccharides are potential candidate biocatalysts for oligosaccharide synthesis. GPs with this linkage specificity are found in two families thus far-glycoside hydrolase family 94 (GH94) and the recently discovered glycoside hydrolase family 149 (GH149). Previously, we reported a crystallographic study of a GH94 laminaribiose phosphorylase with specificity for disaccharides, providing insight into the enzyme's ability to recognize its' sugar substrate/product. In contrast to GH94, characterized GH149 enzymes were shown to have more flexible chain length specificity, with preference for substrate/product with higher degree of polymerization. In order to advance understanding of the specificity of GH149 enzymes, we herein solved X-ray crystallographic structures of GH149 enzyme Pro_7066 in the absence of substrate and in complex with laminarihexaose (G6). The overall domain organization of Pro_7066 is very similar to that of GH94 family enzymes. However, two additional domains flanking its catalytic domain were found only in the GH149 enzyme. Unexpectedly, the G6 complex structure revealed an oligosaccharide surface binding site remote from the catalytic site, which, we suggest, may be associated with substrate targeting. As such, this study reports the first structure of a GH149 phosphorylase enzyme acting on β-(1 → 3)-gluco-oligosaccharides and identifies structural elements that may be involved in defining the specificity of the GH149 enzymes.
Organizational Affiliation:
Department of Biological Chemistry, John Innes Centre, Norwich Research Park, Norwich, UK.