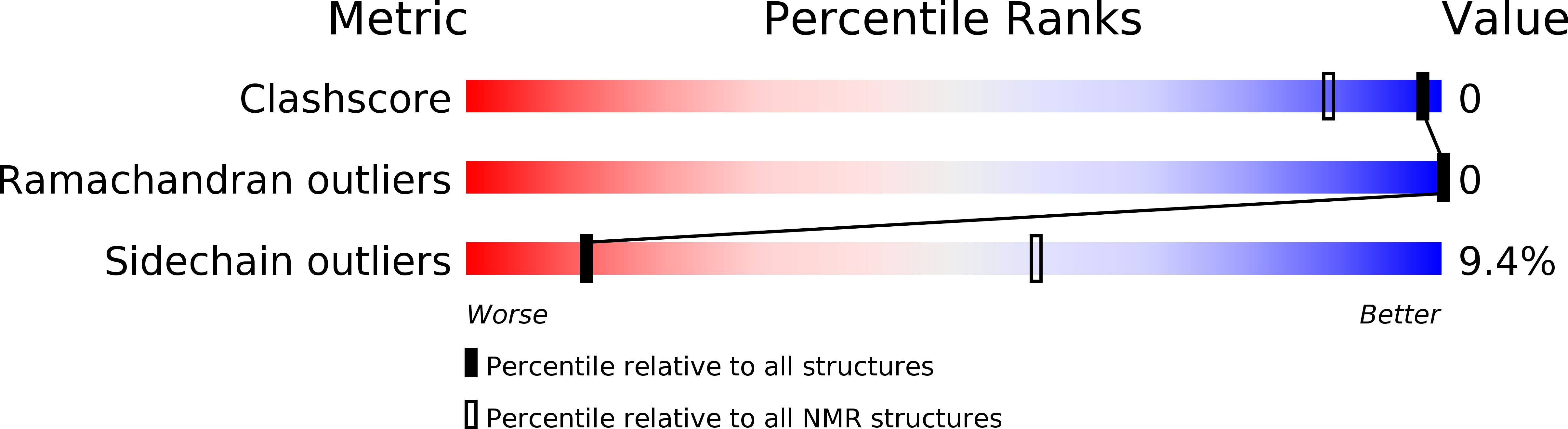An atypical LIR motif within UBA5 (ubiquitin like modifier activating enzyme 5) interacts with GABARAP proteins and mediates membrane localization of UBA5.
Huber, J., Obata, M., Gruber, J., Akutsu, M., Lohr, F., Rogova, N., Guntert, P., Dikic, I., Kirkin, V., Komatsu, M., Dotsch, V., Rogov, V.V.(2020) Autophagy 16: 256-270
- PubMed: 30990354
- DOI: https://doi.org/10.1080/15548627.2019.1606637
- Primary Citation of Related Structures:
6H8C, 6HB9 - PubMed Abstract:
Short linear motifs, known as LC3-interacting regions (LIRs), interact with mactoautophagy/autophagy modifiers (Atg8/LC3/GABARAP proteins) via a conserved universal mechanism. Typically, this includes the occupancy of 2 hydrophobic pockets on the surface of Atg8-family proteins by 2 specific aromatic and hydrophobic residues within the LIR motifs. Here, we describe an alternative mechanism of Atg8-family protein interaction with the non-canonical UBA5 LIR, an E1-like enzyme of the ufmylation pathway that preferentially interacts with GABARAP but not LC3 proteins. By solving the structures of both GABARAP and GABARAPL2 in complex with the UBA5 LIR, we show that in addition to the binding to the 2 canonical hydrophobic pockets (HP1 and HP2), a conserved tryptophan residue N-terminal of the LIR core sequence binds into a novel hydrophobic pocket on the surface of GABARAP proteins, which we term HP0. This mode of action is unique for UBA5 and accompanied by large rearrangements of key residues including the side chains of the gate-keeping K46 and the adjacent K/R47 in GABARAP proteins. Swapping mutations in LC3B and GABARAPL2 revealed that K/R47 is the key residue in the specific binding of GABARAP proteins to UBA5, with synergetic contributions of the composition and dynamics of the loop L3. Finally, we elucidate the physiological relevance of the interaction and show that GABARAP proteins regulate the localization and function of UBA5 on the endoplasmic reticulum membrane in a lipidation-independent manner. Abbreviations: ATG: AuTophaGy-related; EGFP: enhanced green fluorescent protein; GABARAP: GABA-type A receptor-associated protein; ITC: isothermal titration calorimetry; KO: knockout; LIR: LC3-interacting region; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; NMR: nuclear magnetic resonance; RMSD: root-mean-square deviation of atomic positions; TKO: triple knockout; UBA5: ubiquitin like modifier activating enzyme 5.
Organizational Affiliation:
Institute of Biophysical Chemistry and Center for Biomolecular Magnetic Resonance, Goethe University, Frankfurt am Main, Germany.