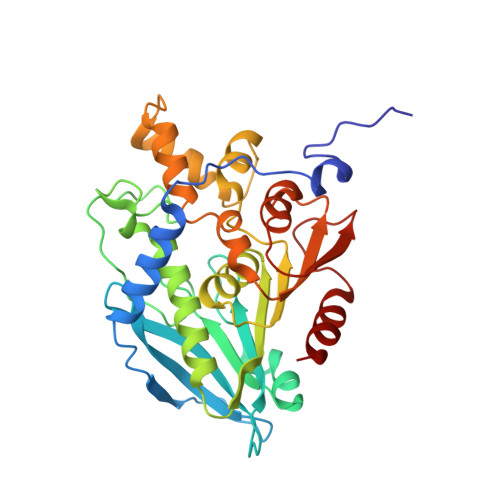
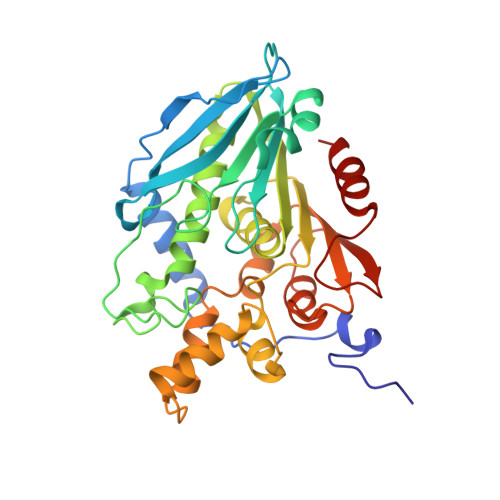
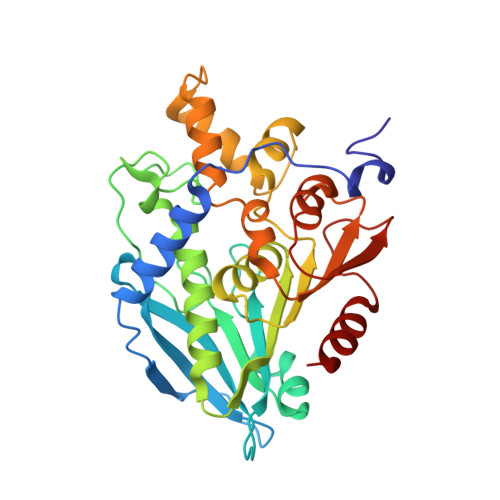
Structural Characterization and Directed Evolution of a Novel Acetyl Xylan Esterase Reveals Thermostability Determinants of the Carbohydrate Esterase 7 Family.
Adesioye, F.A., Makhalanyane, T.P., Vikram, S., Sewell, B.T., Schubert, W.D., Cowan, D.A.(2018) Appl Environ Microbiol 84
- PubMed: 29453256
- DOI: https://doi.org/10.1128/AEM.02695-17
- Primary Citation of Related Structures:
6FKX - PubMed Abstract:
A hot desert hypolith metagenomic DNA sequence data set was screened in silico for genes annotated as acetyl xylan esterases (AcXEs). One of the genes identified encoded an ∼36-kDa protein (Axe1 NaM1 ). The synthesized gene was cloned and expressed, and the resulting protein was purified. NaM1 was optimally active at pH 8.5 and 30°C and functionally stable at salt concentrations of up to 5 M. The specific activity and catalytic efficiency were 488.9 U mg -1 and 3.26 × 10 6 M -1 s -1 , respectively. The crystal structure of wild-type NaM1 was solved at a resolution of 2.03 Å, and a comparison with the structures and models of more thermostable carbohydrate esterase 7 (CE7) family enzymes and variants of NaM1 from a directed evolution experiment suggests that reduced side-chain volume of protein core residues is relevant to the thermal stability of NaM1. Surprisingly, a single point mutation (N96S) not only resulted in a simultaneous improvement in thermal stability and catalytic efficiency but also increased the acyl moiety substrate range of NaM1. IMPORTANCE AcXEs belong to nine carbohydrate esterase families (CE1 to CE7, CE12, and CE16), of which CE7 enzymes possess a unique and narrow specificity for acetylated substrates. All structurally characterized members of this family are moderately to highly thermostable. The crystal structure of a novel, mesophilic CE7 AcXE (Axe1 NaM1 ), from a soil metagenome, provides a basis for comparisons with thermostable CE7 enzymes. Using error-prone PCR and site-directed mutagenesis, we enhanced both the stability and activity of the mesophilic AcXE. With comparative structural analyses, we have also identified possible thermal stability determinants. These are valuable for understanding the thermal stability of enzymes within this family and as a guide for future protein engineering of CE7 and other α/β hydrolase enzymes.
Organizational Affiliation:
Centre for Microbial Ecology and Genomics, Department of Genetics, University of Pretoria, Pretoria, South Africa.