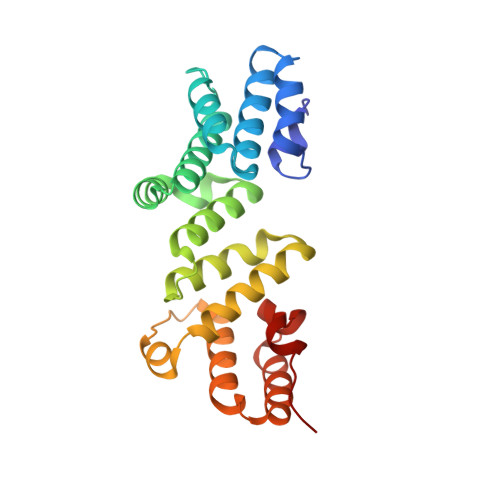
Crystal structures of the amino-terminal domain of LpoA from Escherichia coli and Haemophilus influenzae.
Kelley, A., Vijayalakshmi, J., Saper, M.A.(2019) Acta Crystallogr F Struct Biol Commun 75: 368-376
- PubMed: 31045566
- DOI: https://doi.org/10.1107/S2053230X19004011
- Primary Citation of Related Structures:
6DCJ, 6DR3 - PubMed Abstract:
The bacterial periplasmic protein LpoA is an outer membrane lipoprotein and an activator for the cross-linking activity of PBP1A, a bifunctional peptidoglycan synthase. Previous structures of the amino-terminal (N) domain of LpoA showed it to consist entirely of helices and loops, with at least four tetratricopeptide-like repeats. Although the previously determined orthorhombic crystal structure of the N domain of Haemophilus influenzae LpoA showed a typical curved structure with a concave groove, an NMR structure of the same domain from Escherichia coli was relatively flat. Here, a crystal structure of the N domain of E. coli LpoA was determined to a resolution of 2.1 Å and was found to be more similar to the H. influenzae crystal structure than to the E. coli NMR structure. To provide a quantitative description for these comparisons, the various structures were superimposed pairwise by fitting the first half of each structure to its pairwise partner and then calculating the rotation axis that would optimally superimpose the second half. Differences in both the magnitude of the rotation and the direction of the rotation axis were observed between different pairs of structures. A 1.35 Å resolution structure of a monoclinic crystal form of the N domain of H. influenzae LpoA was also determined. In this structure, the subdomains rotate 10° relative to those in the original orthorhombic H. influenzae crystal structure to further narrow the groove between the subdomains. To accommodate this, a bound chloride ion (in place of sulfate) allowed the closer approach of a helix that forms one side of the groove.
Organizational Affiliation:
Department of Biological Chemistry, University of Michigan, 1150 West Medical Center Drive, Ann Arbor, MI 48109-5606, USA.