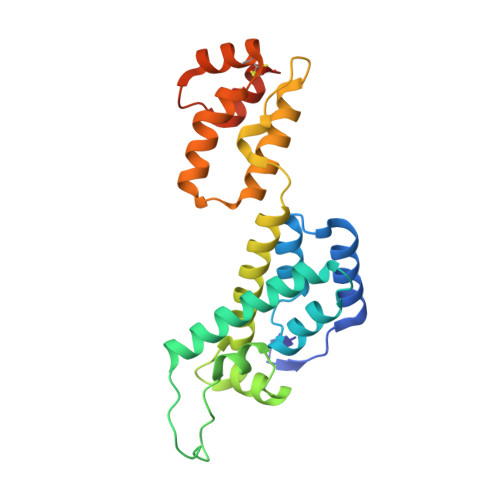
Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization.
Gres, A.T., Kirby, K.A., McFadden, W.M., Du, H., Liu, D., Xu, C., Bryer, A.J., Perilla, J.R., Shi, J., Aiken, C., Fu, X., Zhang, P., Francis, A.C., Melikyan, G.B., Sarafianos, S.G.(2023) Nat Commun 14: 5614-5614
- PubMed: 37699872
- DOI: https://doi.org/10.1038/s41467-023-41197-7
- Primary Citation of Related Structures:
6AY9, 6AYA, 6B2G, 6B2H, 6B2I, 6B2J, 6B2K - PubMed Abstract:
HIV-1 capsid (CA) stability is important for viral replication. E45A and P38A mutations enhance and reduce core stability, thus impairing infectivity. Second-site mutations R132T and T216I rescue infectivity. Capsid lattice stability was studied by solving seven crystal structures (in native background), including P38A, P38A/T216I, E45A, E45A/R132T CA, using molecular dynamics simulations of lattices, cryo-electron microscopy of assemblies, time-resolved imaging of uncoating, biophysical and biochemical characterization of assembly and stability. We report pronounced and subtle, short- and long-range rearrangements: (1) A38 destabilized hexamers by loosening interactions between flanking CA protomers in P38A but not P38A/T216I structures. (2) Two E45A structures showed unexpected stabilizing CA NTD -CA NTD inter-hexamer interactions, variable R18-ring pore sizes, and flipped N-terminal β-hairpin. (3) Altered conformations of E45A a α9-helices compared to WT, E45A/R132T, WT PF74 , WT Nup153 , and WT CPSF6 decreased PF74, CPSF6, and Nup153 binding, and was reversed in E45A/R132T. (4) An environmentally sensitive electrostatic repulsion between E45 and D51 affected lattice stability, flexibility, ion and water permeabilities, electrostatics, and recognition of host factors.
Organizational Affiliation:
C.S. Bond Life Sciences Center, University of Missouri, Columbia, MO, USA.