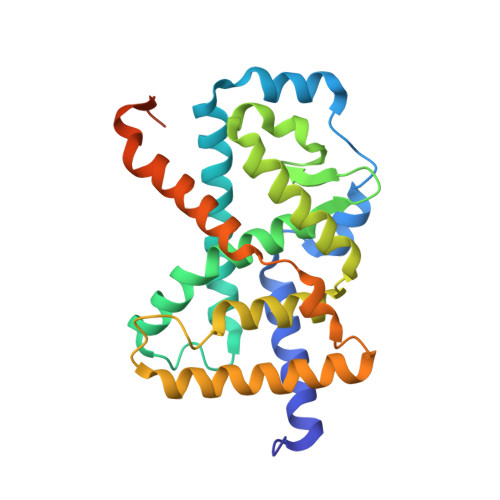
Ternary crystal structure of human ROR gamma ligand-binding-domain, an inhibitor and corepressor peptide provides a new insight into corepressor interaction
Noguchi, M., Nomura, A., Doi, S., Yamaguchi, K., Hirata, K., Shiozaki, M., Maeda, K., Hirashima, S., Kotoku, M., Yamaguchi, T., Katsuda, Y., Crowe, P., Tao, H., Thacher, S., Adachi, T.(2018) Sci Rep 8: 17374-17374
- PubMed: 30478402
- DOI: https://doi.org/10.1038/s41598-018-35783-9
- Primary Citation of Related Structures:
6A22 - PubMed Abstract:
Retinoic acid-related orphan receptor gamma (RORγ) plays pivotal roles in autoimmune diseases by controlling the lineage of interleukin 17 (IL-17)-producing CD4 + T cells (Th17 cells). Structure-based drug design has proven fruitful in the development of inhibitors targeting the ligand binding domain (LBD) of RORγ. Here, we present the crystal structure of a novel RORγ inhibitor co-complex, in the presence of a corepressor (CoR) peptide. This ternary complex with compound T reveals the structural basis for an inhibitory mechanism different from the previously reported inverse agonist. Compared to the inverse agonist, compound T induces about 2 Å shift of helix 5 (H5) backbone and side-chain conformational changes of Met365 on H5. These conformational changes correlate to reduced CoR peptide binding to RORγ-LBD in the presence of compound T, which suggests that the shift of H5 is responsible. This crystal structure analysis will provide useful information for the development of novel and efficacious drugs for autoimmune disorders.
Organizational Affiliation:
Pharmaceutical Frontier Research Laboratories, Central Pharmaceutical Research Institute, Japan Tobacco Inc., 1-13-2, Fukuura, Kanazawa-Ku, Yokohama, Kanagawa, 236-0004, Japan. masato.noguchi@jt.com.