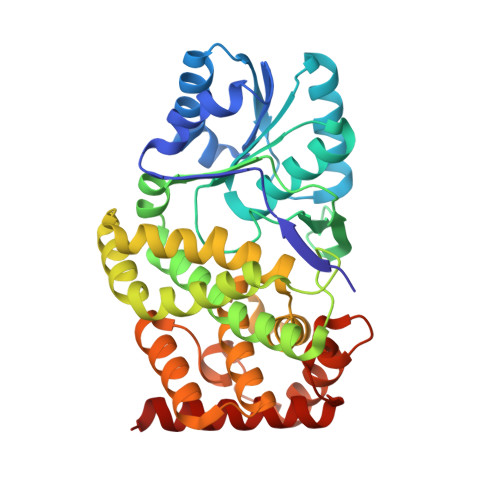
Seeing but not believing: the structure of glycerol dehydrogenase initially assumed to be the structure of a survival protein from Salmonella typhimurium
Hatti, K., Mathiharan, Y.K., Srinivasan, N., Murthy, M.R.N.(2017) Acta Crystallogr D Biol Crystallogr 73: 609-617