Structural insights into the ubiquitin recognition by OPTN (optineurin) and its regulation by TBK1-mediated phosphorylation.
Li, F., Xu, D., Wang, Y., Zhou, Z., Liu, J., Hu, S., Gong, Y., Yuan, J., Pan, L.(2018) Autophagy 14: 66-79
- PubMed: 29394115
- DOI: https://doi.org/10.1080/15548627.2017.1391970
- Primary Citation of Related Structures:
5WQ4 - PubMed Abstract:
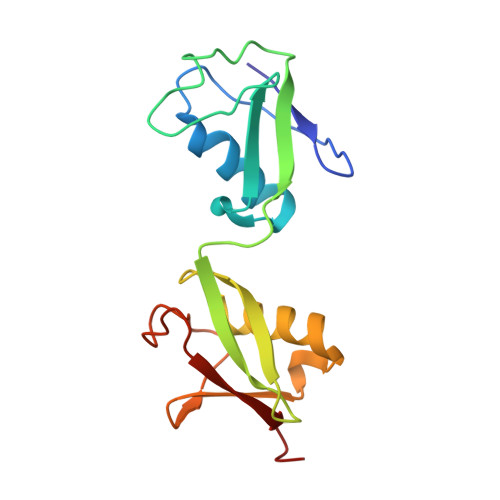
OPTN (optineurin), a ubiquitin-binding scaffold protein, functions as an important macroautophagy/autophagy receptor in selective autophagy processes. Mutations in OPTN have been linked with human neurodegenerative diseases including ALS and glaucoma. However, the mechanistic basis underlying the recognition of ubiquitin by OPTN and its regulation by TBK1-mediated phosphorylation are still elusive. Here, we demonstrate that the UBAN domain of OPTN preferentially recognizes linear ubiquitin chain and forms an asymmetric 2:1 stoichiometry complex with the linear diubiquitin. In addition, our results provide new mechanistic insights into how phosphorylation of UBAN would regulate the ubiquitin-binding ability of OPTN and how disease-associated mutations in the OPTN UBAN domain disrupt its interaction with ubiquitin. Finally, we show that defects in ubiquitin-binding may affect the recruitment of OPTN to linear ubiquitin-decorated mutant Huntington protein aggregates. Taken together, our findings clarify the interaction mode between UBAN and linear ubiquitin chain in general, and expand our knowledge of the molecular mechanism of ubiquitin-decorated substrates recognition by OPTN as well as the pathogenesis of neurodegenerative diseases caused by OPTN mutations.
Organizational Affiliation:
a State Key Laboratory of Bioorganic and Natural Products Chemistry , Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences , Shanghai , China.