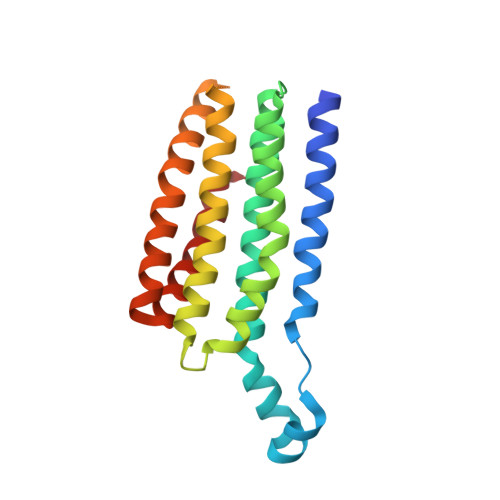
The lysosomal potassium channel TMEM175 adopts a novel tetrameric architecture.
Lee, C., Guo, J., Zeng, W., Kim, S., She, J., Cang, C., Ren, D., Jiang, Y.(2017) Nature 547: 472-475
- PubMed: 28723891
- DOI: https://doi.org/10.1038/nature23269
- Primary Citation of Related Structures:
5VRE - PubMed Abstract:
TMEM175 is a lysosomal K + channel that is important for maintaining the membrane potential and pH stability in lysosomes. It contains two homologous copies of a six-transmembrane-helix (6-TM) domain, which has no sequence homology to the canonical tetrameric K + channels and lacks the TVGYG selectivity filter motif found in these channels. The prokaryotic TMEM175 channel, which is present in a subset of bacteria and archaea, contains only a single 6-TM domain and functions as a tetramer. Here, we present the crystal structure of a prokaryotic TMEM175 channel from Chamaesiphon minutus, CmTMEM175, the architecture of which represents a completely different fold from that of canonical K + channels. All six transmembrane helices of CmTMEM175 are tightly packed within each subunit without undergoing domain swapping. The highly conserved TM1 helix acts as the pore-lining inner helix, creating an hourglass-shaped ion permeation pathway in the channel tetramer. Three layers of hydrophobic residues on the carboxy-terminal half of the TM1 helices form a bottleneck along the ion conduction pathway and serve as the selectivity filter of the channel. Mutagenesis analysis suggests that the first layer of the highly conserved isoleucine residues in the filter is primarily responsible for channel selectivity. Thus, the structure of CmTMEM175 represents a novel architecture of a tetrameric cation channel whose ion selectivity mechanism appears to be distinct from that of the classical K + channel family.
Organizational Affiliation:
Department of Physiology and Department of Biophysics, University of Texas Southwestern Medical Center, Dallas, Texas 75390-9040, USA.