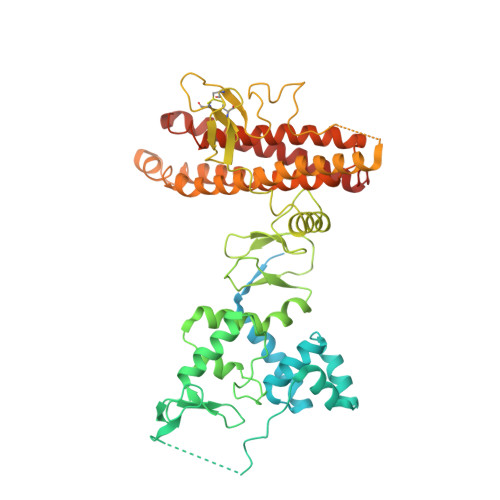
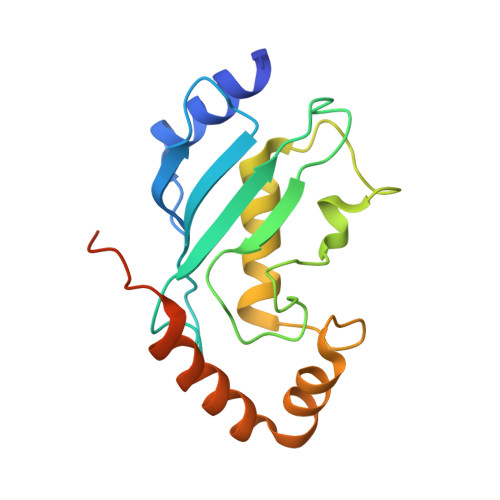
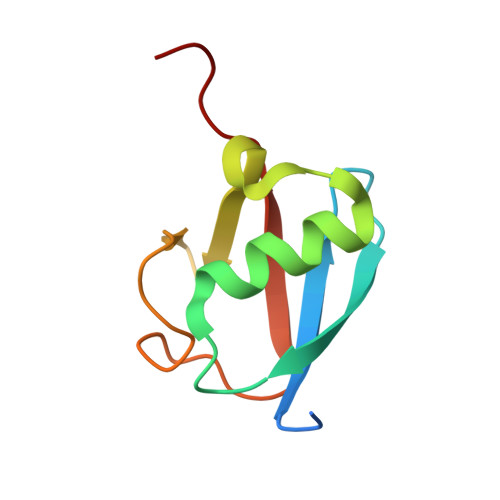
Structural insights into the mechanism and E2 specificity of the RBR E3 ubiquitin ligase HHARI.
Yuan, L., Lv, Z., Atkison, J.H., Olsen, S.K.(2017) Nat Commun 8: 211-211
- PubMed: 28790309
- DOI: https://doi.org/10.1038/s41467-017-00272-6
- Primary Citation of Related Structures:
5TTE - PubMed Abstract:
RING-in-between-RING (RBR) ubiquitin (Ub) E3 ligases function with Ub E2s through a RING/HECT hybrid mechanism to conjugate Ub to target proteins. Here, we report the crystal structure of the RBR E3, HHARI, in complex with a UbcH7 ~ Ub thioester mimetic which reveals the molecular basis for the specificity of this cognate E2/RBR E3 pair. The structure also reveals mechanistically important conformational changes in the RING1 and UBA-like domains of HHARI that accompany UbcH7 ~ Ub binding and provides a molecular basis by which HHARI recruits E2 ~ Ub in an 'open' conformation. In addition to optimally functioning with an E2 that solely performs transthiolation, our data suggests that HHARI prevents spurious discharge of Ub from E2 to lysine residues by: (1) harboring structural elements that block E2 ~ Ub from adopting a 'closed' conformation and (2) participating in contacts to ubiquitin that promote an open E2 ~ Ub conformation.HHARI is a RING-in-between-RING (RBR) ubiquitin (Ub) E3 ligase. Here the authors present the crystal structure of HHARI with the UbcH7 ~ Ub thioester intermediate mimetic, which reveals that HHARI binds this E2 ~ Ub in an open conformation and explains the specificity of this cognate RBR E3/E2 pair.
Organizational Affiliation:
Department of Biochemistry & Molecular Biology, Medical University of South Carolina, Charleston, SC, 29425, USA.