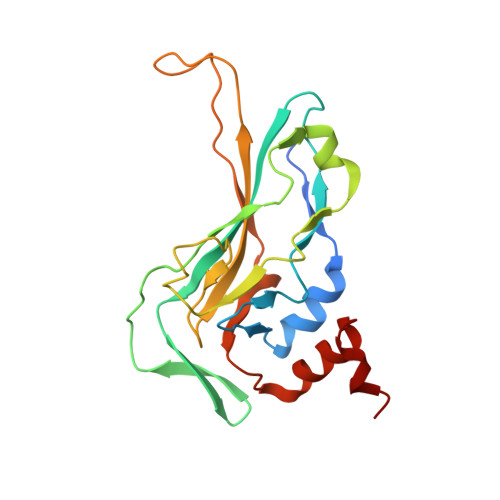
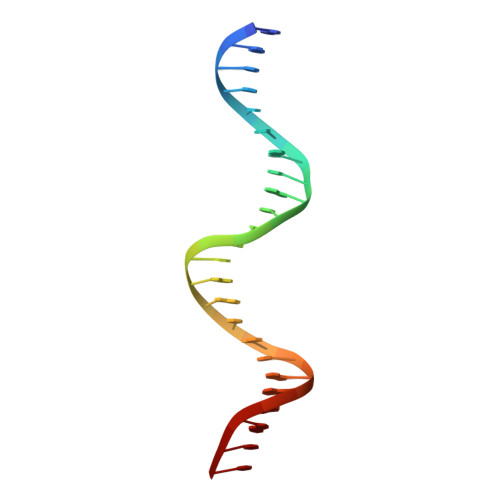
Crystal structure of the DNA binding domain of the transcription factor T-bet suggests simultaneous recognition of distant genome sites.
Liu, C.F., Brandt, G.S., Hoang, Q.Q., Naumova, N., Lazarevic, V., Hwang, E.S., Dekker, J., Glimcher, L.H., Ringe, D., Petsko, G.A.(2016) Proc Natl Acad Sci U S A 113: E6572-E6581
- PubMed: 27791029
- DOI: https://doi.org/10.1073/pnas.1613914113
- Primary Citation of Related Structures:
5T1J - PubMed Abstract:
The transcription factor T-bet (Tbox protein expressed in T cells) is one of the master regulators of both the innate and adaptive immune responses. It plays a central role in T-cell lineage commitment, where it controls the T H 1 response, and in gene regulation in plasma B-cells and dendritic cells. T-bet is a member of the Tbox family of transcription factors; however, T-bet coordinately regulates the expression of many more genes than other Tbox proteins. A central unresolved question is how T-bet is able to simultaneously recognize distant Tbox binding sites, which may be located thousands of base pairs away. We have determined the crystal structure of the Tbox DNA binding domain (DBD) of T-bet in complex with a palindromic DNA. The structure shows a quaternary structure in which the T-bet dimer has its DNA binding regions splayed far apart, making it impossible for a single dimer to bind both sites of the DNA palindrome. In contrast to most other Tbox proteins, a single T-bet DBD dimer binds simultaneously to identical half-sites on two independent DNA. A fluorescence-based assay confirms that T-bet dimers are able to bring two independent DNA molecules into close juxtaposition. Furthermore, chromosome conformation capture assays confirm that T-bet functions in the direct formation of chromatin loops in vitro and in vivo. The data are consistent with a looping/synapsing model for transcriptional regulation by T-bet in which a single dimer of the transcription factor can recognize and coalesce distinct genetic elements, either a promoter plus a distant regulatory element, or promoters on two different genes.
Organizational Affiliation:
Appel Alzheimer's Disease Research Institute, Brain and Mind Research Institute, Weill Cornell Medical College, New York, NY 10021; Rosenstiel Basic Medical Sciences Research Center, Brandeis University, Waltham, MA 02454; cel2010@med.cornell.edu gpetsko@med.cornell.edu.