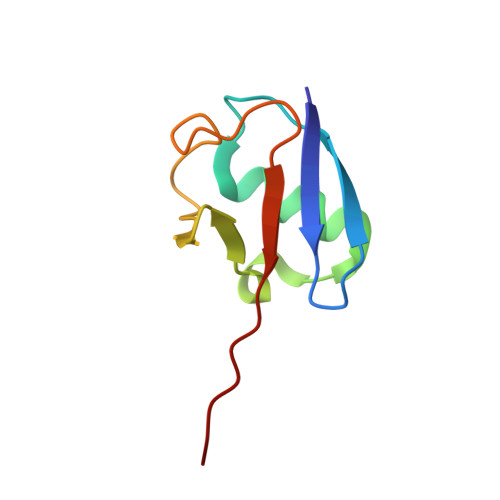
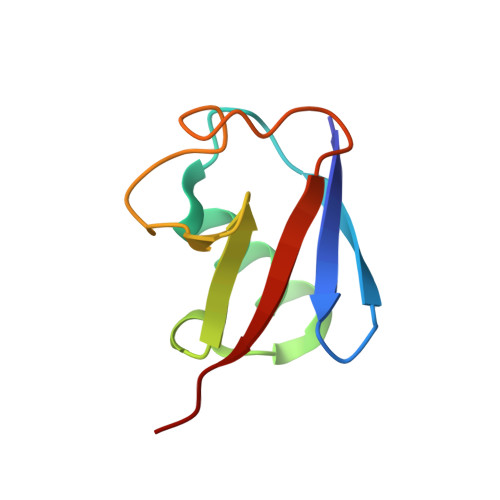
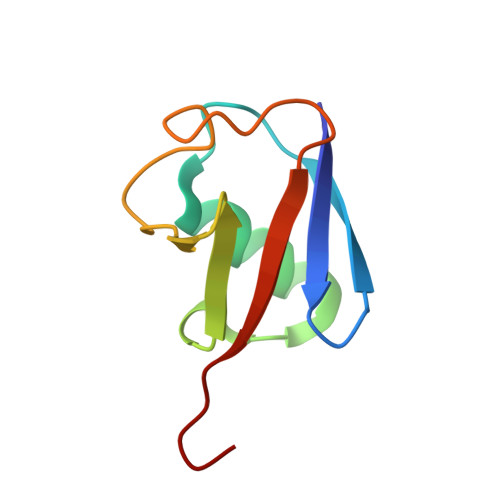
The Crystal Structure and Conformations of an Unbranched Mixed Tri-Ubiquitin Chain Containing K48 and K63 Linkages.
Padala, P., Soudah, N., Giladi, M., Haitin, Y., Isupov, M.N., Wiener, R.(2017) J Mol Biol 429: 3801-3813
- PubMed: 29111344
- DOI: https://doi.org/10.1016/j.jmb.2017.10.027
- Primary Citation of Related Structures:
5O44 - PubMed Abstract:
The ability of ubiquitin to function in a wide range of cellular processes is ascribed to its capacity to cause a diverse spectrum of modifications. While a target protein can be modified with monoubiquitin, it can also be modified with ubiquitin chains. The latter include seven types of homotypic chains as well as mixed ubiquitin chains. In a mixed chain, not all the isopeptide bonds are restricted to a specific lysine of ubiquitin, resulting in a chain possessing more than one type of linkage. While structural characterization of homotypic chains has been well elucidated, less is known about mixed chains. Here we present the crystal structure of a mixed tri-ubiquitin chain at 3.1-Å resolution. In the structure, the proximal ubiquitin is connected to the middle ubiquitin via K48 and these two ubiquitins adopt a compact structure as observed in K48 di-ubiquitin. The middle ubiquitin links to the distal ubiquitin via its K63 and these ubiquitins adopt two conformations, suggesting a flexible structure. Using small-angle X-ray scattering, we unexpectedly found differences between the conformational ensembles of the above tri-ubiquitin chains and chains possessing the same linkages but in the reverse order. In addition, cleavage of the K48 linkage by DUB is faster if this linkage is at the distal end. Taken together, our results suggest that in mixed chains, not only the type of the linkages but also their sequence determine the structural and functional properties of the chain.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, the Institute for Medical Research Israel-Canada, Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel.