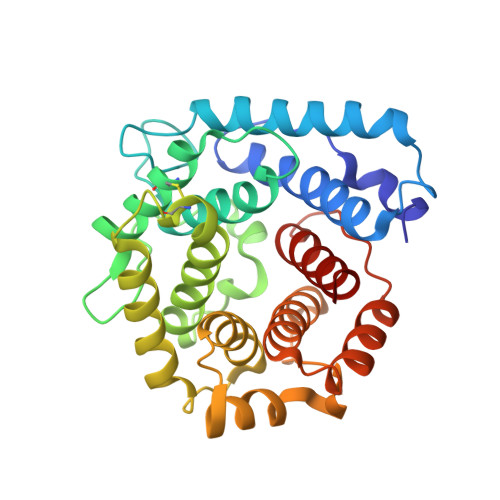
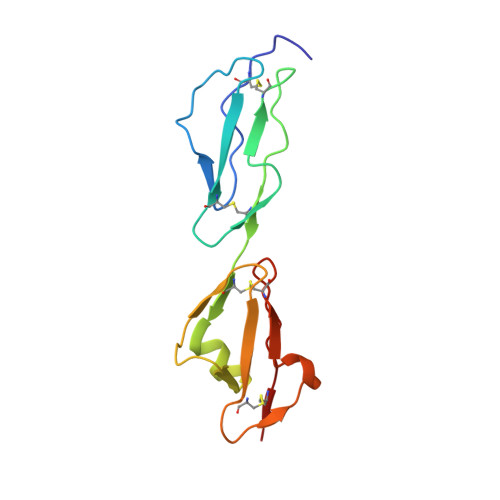
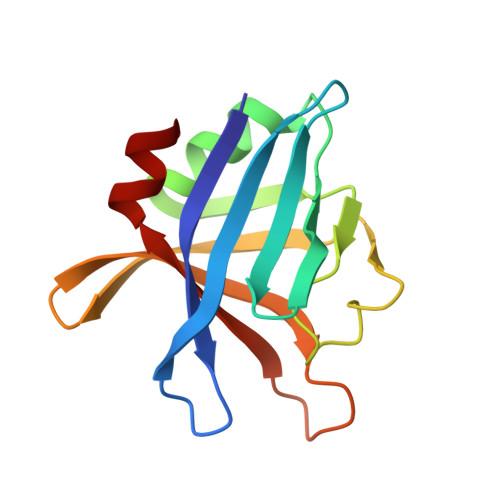
Crystal structure of a tripartite complex between C3dg, C-terminal domains of factor H and OspE of Borrelia burgdorferi.
Kolodziejczyk, R., Mikula, K.M., Kotila, T., Postis, V.L.G., Jokiranta, T.S., Goldman, A., Meri, T.(2017) PLoS One 12: e0188127-e0188127
- PubMed: 29190743
- DOI: https://doi.org/10.1371/journal.pone.0188127
- PubMed Abstract:
Complement is an important part of innate immunity. The alternative pathway of complement is activated when the main opsonin, C3b coats non-protected surfaces leading to opsonisation, phagocytosis and cell lysis. The alternative pathway is tightly controlled to prevent autoactivation towards host cells. The main regulator of the alternative pathway is factor H (FH), a soluble glycoprotein that terminates complement activation in multiple ways. FH recognizes host cell surfaces via domains 19-20 (FH19-20). All microbes including Borrelia burgdorferi, the causative agent of Lyme borreliosis, must evade complement activation to allow the infectious agent to survive in its host. One major mechanism that Borrelia uses is to recruit FH from host. Several outer surface proteins (Osp) have been described to bind FH via the C-terminus, and OspE is one of them. Here we report the structure of the tripartite complex formed by OspE, FH19-20 and C3dg at 3.18 Å, showing that OspE and C3dg can bind simultaneously to FH19-20. This verifies that FH19-20 interacts via the "common microbial binding site" on domain 20 with OspE and simultaneously and independently via domain 19 with C3dg. The spatial organization of the tripartite complex explains how OspE on the bacterial surface binds FH19-20, leaving FH fully available to protect the bacteria against complement. Additionally, formation of tripartite complex between FH, microbial protein and C3dg might enable enhanced protection, particularly on those regions on the bacteria where previous complement activation led to deposition of C3d. This might be especially important for slow-growing bacteria that cause chronic disease like Borrelia burgdorferi.
Organizational Affiliation:
Division of Biochemistry, Department of Biosciences, University of Helsinki, Helsinki, Finland.