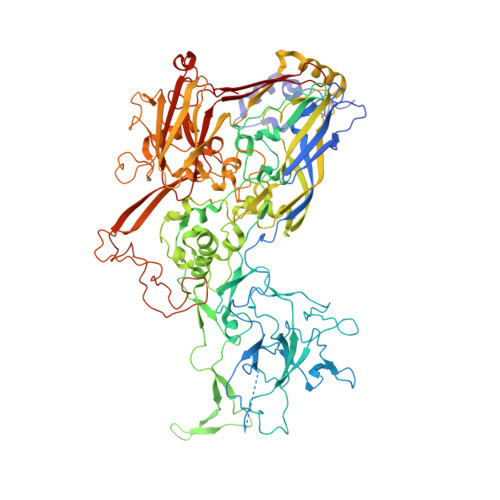
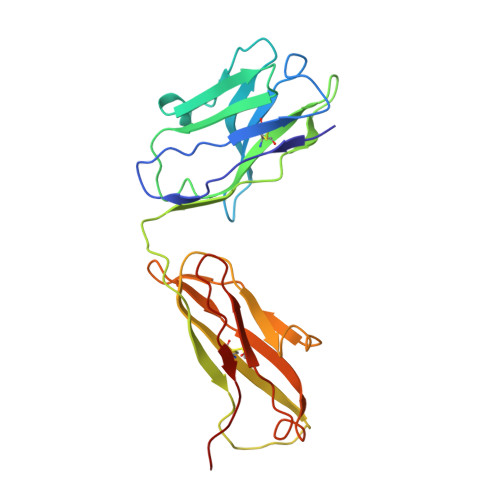
Antibody-antigen kinetics constrain intracellular humoral immunity.
Bottermann, M., Lode, H.E., Watkinson, R.E., Foss, S., Sandlie, I., Andersen, J.T., James, L.C.(2016) Sci Rep 6: 37457-37457
- PubMed: 27881870
- DOI: https://doi.org/10.1038/srep37457
- Primary Citation of Related Structures:
5LDN - PubMed Abstract:
During infection with non-enveloped viruses, antibodies stimulate immunity from inside cells by activating the cytosolic Fc receptor TRIM21. This intracellular humoral response relies on opsonized viral particles reaching the cytosol intact but the antigenic and kinetic constraints involved are unknown. We have solved the structure of a potent TRIM21-dependent neutralizing antibody in complex with human adenovirus 5 hexon and show how these properties influence immune activity. Structure-guided mutagenesis was used to generate antibodies with 20,000-fold variation in affinity, on-rates that differ by ~50-fold and off-rates by >175-fold. Characterization of these variants during infection revealed that TRIM21-dependent neutralization and NFκB activation was largely unaffected by on-rate kinetics. In contrast, TRIM21 antiviral activity was exquisitely dependent upon off-rate, with sub-μM affinity antibodies nevertheless unable to stimulate signaling because of fast dissociation kinetics. These results define the antibody properties required to elicit an efficient intracellular immune response during viral infection.
Organizational Affiliation:
Medical Research Council Laboratory of Molecular Biology, Division of Protein and Nucleic Acid Chemistry, Francis Crick Avenue, Cambridge, CB2 0QH, United Kingdom.