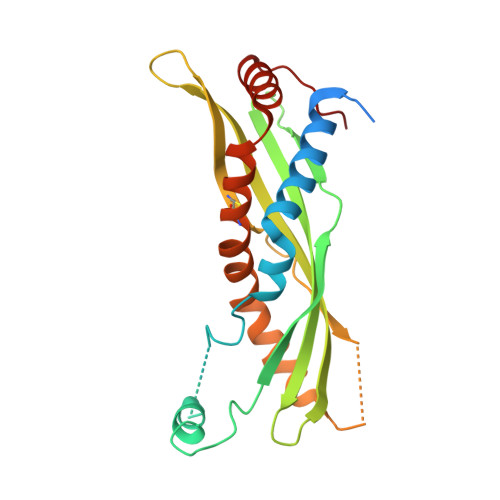
Structural Features Essential to the Antimicrobial Functions of Human SPLUNC1.
Walton, W.G., Ahmad, S., Little, M.S., Kim, C.S., Tyrrell, J., Lin, Q., Di, Y.P., Tarran, R., Redinbo, M.R.(2016) Biochemistry 55: 2979-2991
- PubMed: 27145151
- DOI: https://doi.org/10.1021/acs.biochem.6b00271
- Primary Citation of Related Structures:
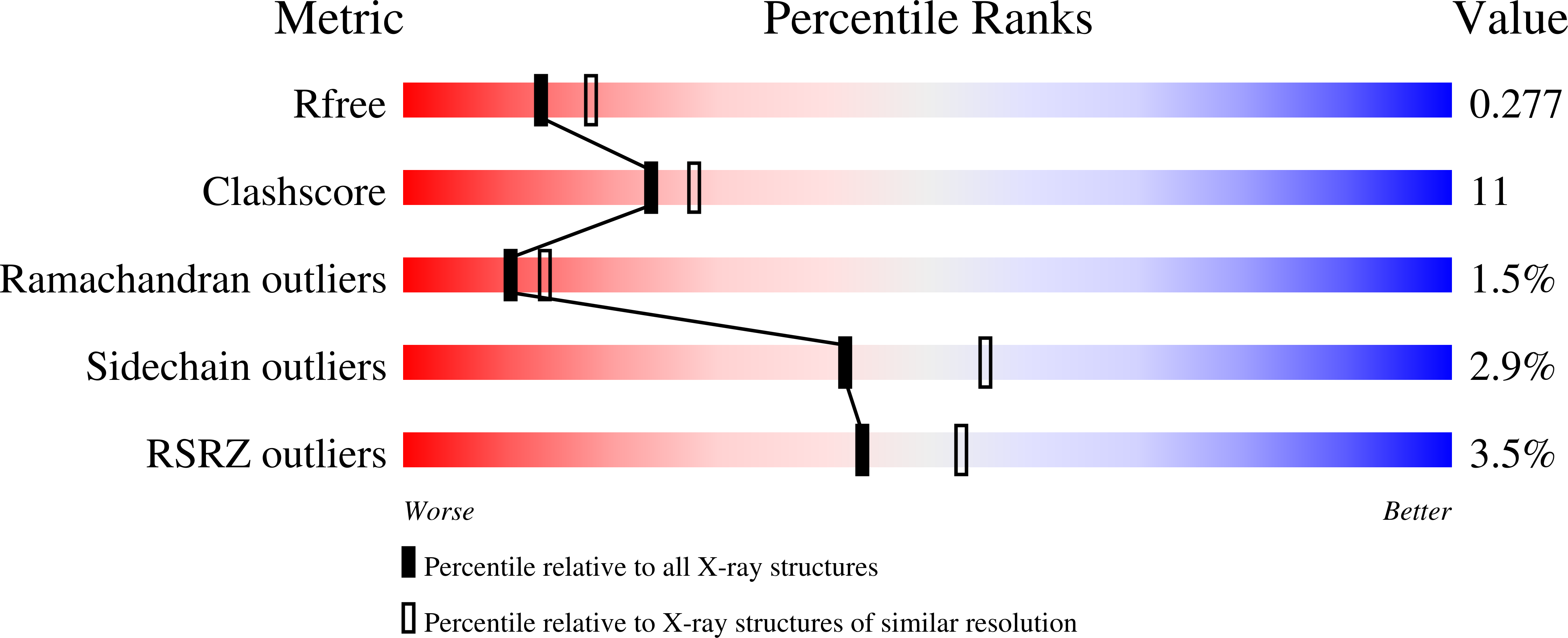
5I7J, 5I7K, 5I7L - PubMed Abstract:
SPLUNC1 is an abundantly secreted innate immune protein in the mammalian respiratory tract that exerts bacteriostatic and antibiofilm effects, binds to lipopolysaccharide (LPS), and acts as a fluid-spreading surfactant. Here, we unravel the structural elements essential for the surfactant and antimicrobial functions of human SPLUNC1 (short palate lung nasal epithelial clone 1). A unique α-helix (α4) that extends from the body of SPLUNC1 is required for the bacteriostatic, surfactant, and LPS binding activities of this protein. Indeed, we find that mutation of just four leucine residues within this helical motif to alanine is sufficient to significantly inhibit the fluid spreading abilities of SPLUNC1, as well as its bacteriostatic actions against Gram-negative pathogens Burkholderia cenocepacia and Pseudomonas aeruginosa. Conformational flexibility in the body of SPLUNC1 is also involved in the bacteriostatic, surfactant, and LPS binding functions of the protein as revealed by disulfide mutants introduced into SPLUNC1. In addition, SPLUNC1 exerts antibiofilm effects against Gram-negative bacteria, although α4 is not involved in this activity. Interestingly, though, the introduction of surface electrostatic mutations away from α4 based on the unique dolphin SPLUNC1 sequence, and confirmed by crystal structure, is shown to impart antibiofilm activity against Staphylococcus aureus, the first SPLUNC1-dependent effect against a Gram-positive bacterium reported to date. Together, these data pinpoint SPLUNC1 structural motifs required for the antimicrobial and surfactant actions of this protective human protein.
Organizational Affiliation:
Departments of Chemistry, Biochemistry, and Microbiology, University of North Carolina , 4350 Genome Sciences Building, Chapel Hill, North Carolina 27599-3290, United States.