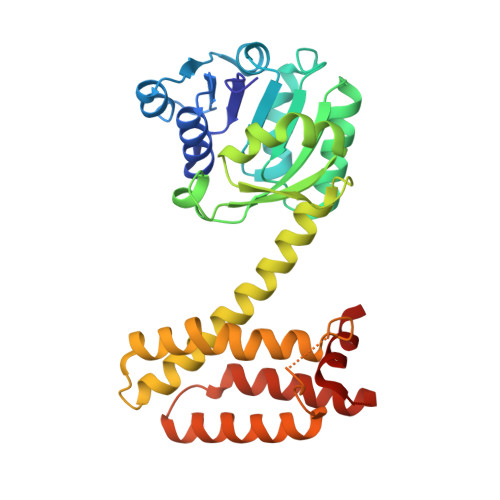
Stereoselectivity and Structural Characterization of an Imine Reductase (Ired) from Amycolatopsis Orientalis
Aleku, G.A., Man, H., France, S.P., Leipold, F., Hussain, S., Toca-Gonzalez, L., Marchington, R., Hart, S., Turkenburg, J.P., Grogan, G., Turner, N.J.(2016) ACS Catal 6: 3880