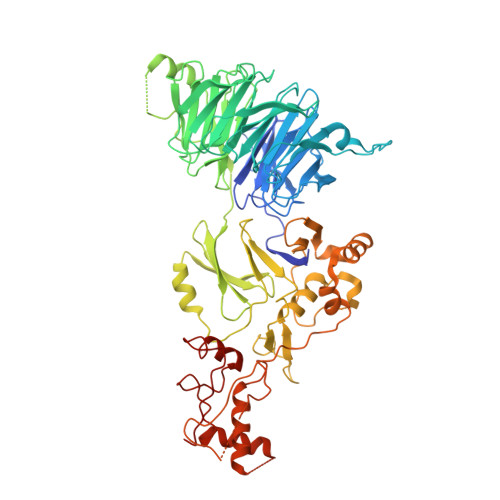
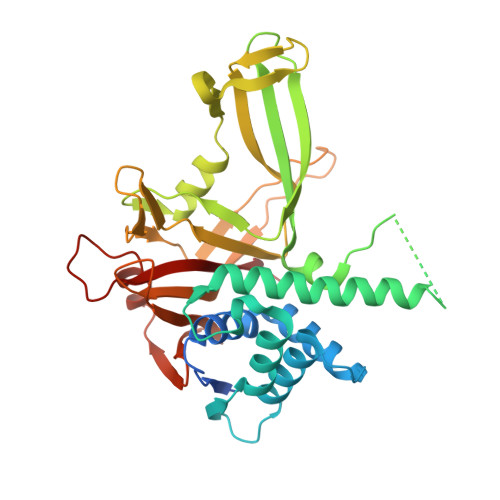
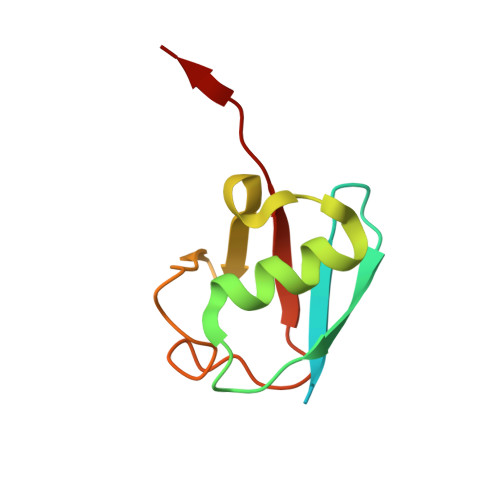
Structural Insights into WD-Repeat 48 Activation of Ubiquitin-Specific Protease 46.
Yin, J., Schoeffler, A.J., Wickliffe, K., Newton, K., Starovasnik, M.A., Dueber, E.C., Harris, S.F.(2015) Structure 23: 2043-2054
- PubMed: 26388029
- DOI: https://doi.org/10.1016/j.str.2015.08.010
- PubMed Abstract:
Protein ubiquitination patterns are an important component of cellular signaling. The WD-repeat protein WDR48 (USP1-associated factor UAF-1) stimulates activity of ubiquitin-specific proteases USP1, USP12, and USP46. To understand how WDR48 exerts its effect on the USP scaffold, we determined structures of the ternary WDR48:USP46:ubiquitin complex. WDR48 interacts with the USP46 fingers subdomain via a relatively small, highly polar surface on the top center of the WDR48 β propeller. In addition, WDR48 has a novel ancillary domain and a C-terminal SUMO-like domain encircling the USP46-bound ubiquitin. Mutation of residues involved in the WDR48:USP46 interaction abrogated both binding and deubiquitinase activity of the complex. An analogous mutation in USP1 similarly blocked WDR48-dependent activation. Our data suggest a possible mechanism of deubiquitinase stimulation via stabilization and prolonged residence time of substrate. The unprecedented mode of interaction between the USP fingers domain and the WD-repeat β propeller serves as a prototypical example for this family of deubiquitinases.
Organizational Affiliation:
Department of Structural Biology, Genentech, Inc., 1 DNA Way, South San Francisco, CA 94080, USA.