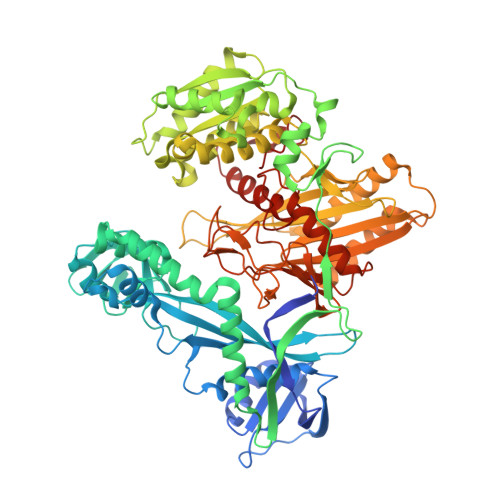
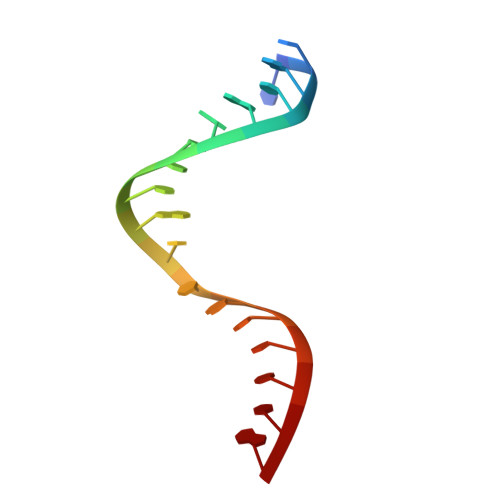
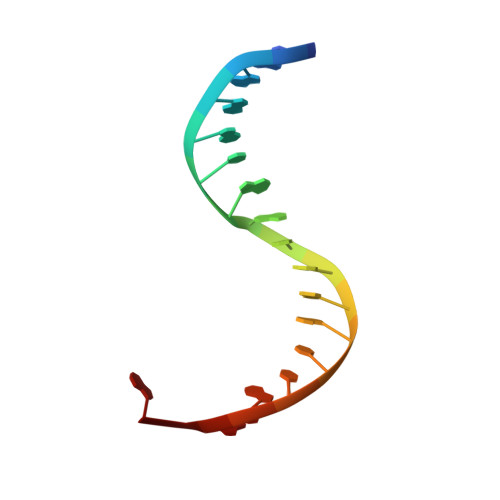
Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute.
Miyoshi, T., Ito, K., Murakami, R., Uchiumi, T.(2016) Nat Commun 7: 11846-11846
- PubMed: 27325485
- DOI: https://doi.org/10.1038/ncomms11846
- Primary Citation of Related Structures:
5AWH - PubMed Abstract:
Argonaute proteins are key players in the gene silencing mechanisms mediated by small nucleic acids in all domains of life from bacteria to eukaryotes. However, little is known about the Argonaute protein that recognizes guide RNA/target DNA. Here, we determine the 2 Å crystal structure of Rhodobacter sphaeroides Argonaute (RsAgo) in a complex with 18-nucleotide guide RNA and its complementary target DNA. The heteroduplex maintains Watson-Crick base-pairing even in the 3'-region of the guide RNA between the N-terminal and PIWI domains, suggesting a recognition mode by RsAgo for stable interaction with the target strand. In addition, the MID/PIWI interface of RsAgo has a system that specifically recognizes the 5' base-U of the guide RNA, and the duplex-recognition loop of the PAZ domain is important for the DNA silencing activity. Furthermore, we show that Argonaute discriminates the nucleic acid type (RNA/DNA) by recognition of the duplex structure of the seed region.
Organizational Affiliation:
Center for Transdisciplinary Research, Niigata University, 8050 Ikarashi 2-no-cho, Nishi-ku, Niigata 950-2181, Japan.