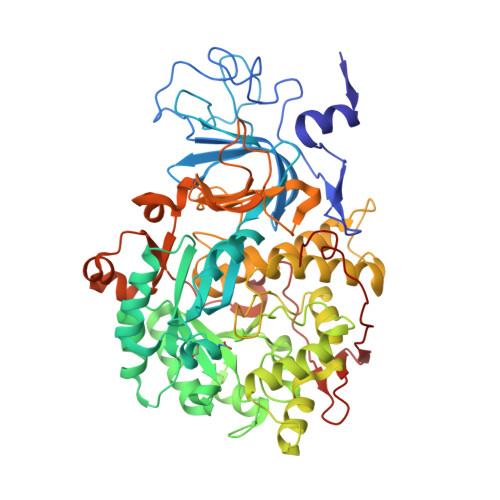
Kinetic and Structural Studies Reveal a Unique Binding Mode of Sulfite to the Nickel Center in Urease.
Mazzei, L., Cianci, M., Benini, S., Bertini, L., Musiani, F., Ciurli, S.(2015) J Inorg Biochem 154: 42
- PubMed: 26580226
- DOI: https://doi.org/10.1016/j.jinorgbio.2015.11.003
- Primary Citation of Related Structures:
5A6T - PubMed Abstract:
Urease is the most efficient enzyme known to date, and catalyzes the hydrolysis of urea using two Ni(II) ions in the active site. Urease is a virulence factor in several human pathogens, while causing severe environmental and agronomic problems. Sporosarcina pasteurii urease has been used extensively in the structural characterization of the enzyme. Sodium sulfite has been widely used as a preservative in urease solutions to prevent oxygen-induced oxidation, but its role as an inhibitor has also been suggested. In the present study, isothermal titration microcalorimetry was used to establish sulfite as a competitive inhibitor for S. pasteurii urease, with an inhibition constant of 0.19mM at pH7. The structure of the urease-sulfite complex, determined at 1.65Å resolution, shows the inhibitor bound to the dinuclear Ni(II) center of urease in a tridentate mode involving bonds between the two Ni(II) ions in the active site and all three oxygen atoms of the inhibitor, supporting the observed competitive inhibition kinetics. This coordination mode of sulfite has never been observed, either in proteins or in small molecule complexes, and could inspire synthetic coordination chemists as well as biochemists to develop urease inhibitors based on this chemical moiety.
Organizational Affiliation:
Laboratory of Bioinorganic Chemistry, Department of Pharmacy and Biotechnology, University of Bologna, Via Giuseppe Fanin 40, I-40138, Bologna, Italy.