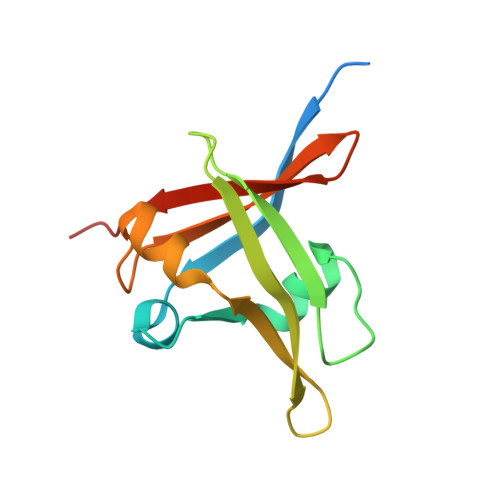
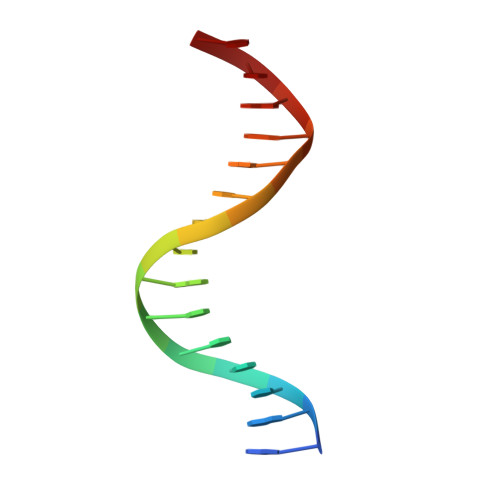
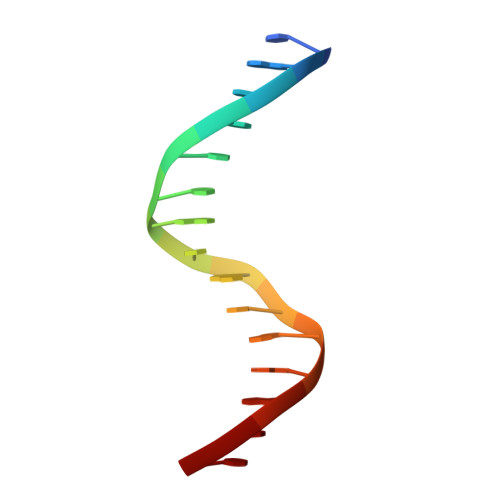
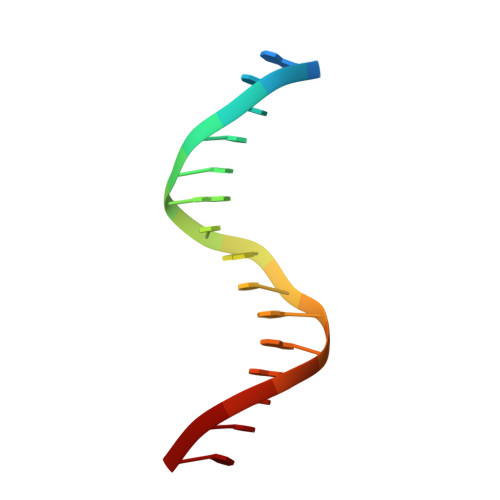
Structural insight into the role of VAL1 B3 domain for targeting to FLC locus in Arabidopsis thaliana.
Wu, B.X., Zhang, M.M., Su, S.C., Liu, H.H., Gan, J.H., Ma, J.B.(2018) Biochem Biophys Res Commun
- PubMed: 29733847
- DOI: https://doi.org/10.1016/j.bbrc.2018.05.002
- Primary Citation of Related Structures:
5YZY, 5YZZ, 5Z00 - PubMed Abstract:
Vernalization is a pivotal stage for some plants involving many epigenetic changes during cold exposure. In Arabidopsis, an essential step in vernalization for further flowering is successful silence the potent floral repressor Flowering Locus C (FLC) by repressing histone mark. AtVal1 is a multi-function protein containing five domains that participate into many recognition processes and is validated to recruit the repress histone modifier PHD-PRC2 complex and interact with components of the ASAP complex target to the FLC nucleation region through recognizing a cis element known as CME (cold memory element) by its plant-specific B3 domain. Here, we determine the crystal structure of the B3 domain in complex with Sph/RY motif in CME. Our structural analysis reveals the specific DNA recognition by B3 domain, combined with our in vitro experiments, we provide the structural insight into the important implication of AtVAL1-B3 domain in flowering process.
Organizational Affiliation:
State Key Laboratory of Genetic Engineering, Collaborative Innovation Centre of Genetics and Development, Department of Biochemistry, Institute of Plant Biology, School of Life Sciences, Fudan University, Shanghai 200438, China. Electronic address: bxwu15@fudan.edu.cn.