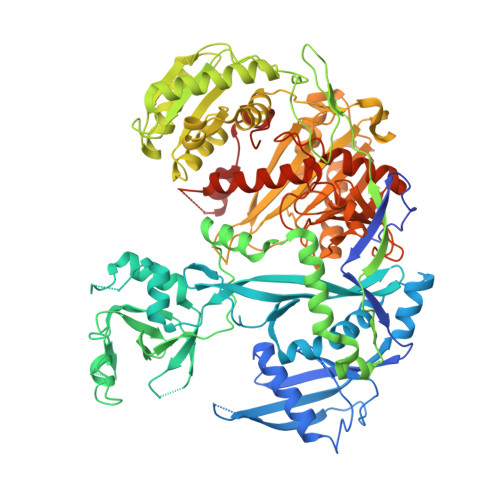
Multivalent Recruitment of Human Argonaute by GW182.
Elkayam, E., Faehnle, C.R., Morales, M., Sun, J., Li, H., Joshua-Tor, L.(2017) Mol Cell 67: 646-658.e3
- PubMed: 28781232
- DOI: https://doi.org/10.1016/j.molcel.2017.07.007
- PubMed Abstract:
In miRNA-mediated gene silencing, the physical interaction between human Argonaute (hAgo) and GW182 (hGW182) is essential for facilitating the downstream silencing of the targeted mRNA. GW182 can interact with hAgo via three of the GW/WG repeats in its Argonaute-binding domain: motif-1, motif-2, and the hook motif. The structure of hAgo1 in complex with the hook motif of hGW182 reveals a "gate"-like interaction that is critical for GW182 docking into one of hAgo1's tryptophan-binding pockets. We show that hAgo1 and hAgo2 have a single GW182-binding site and that miRNA binding increases hAgo's affinity to GW182. With target binding occurring rapidly, this ensures that only mature RISC would be recruited for silencing. Finally, we show that hGW182 can recruit up to three copies of hAgo via its three GW motifs. This may explain the observed cooperativity in miRNA-mediated gene silencing.
Organizational Affiliation:
Keck Structural Biology Laboratory, Howard Hughes Medical Institute, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 11724, USA.