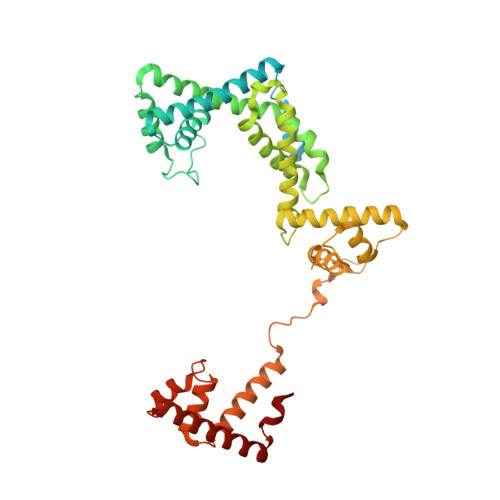
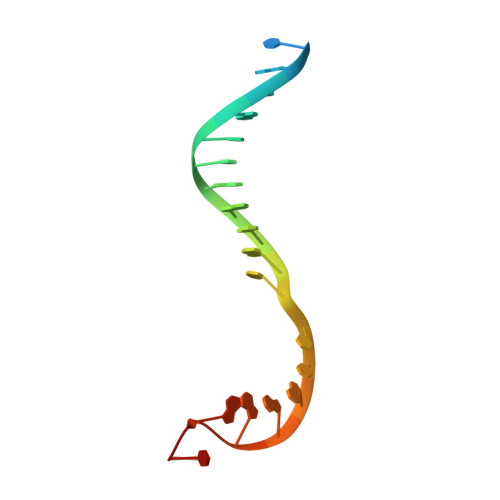
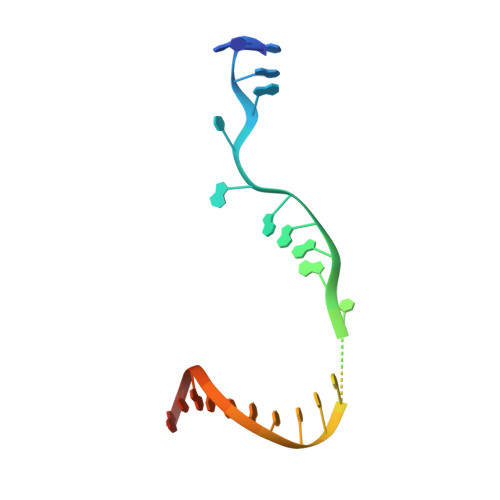
X-ray crystal structure of a reiterative transcription complex reveals an atypical RNA extension pathway.
Murakami, K.S., Shin, Y., Turnbough, C.L., Molodtsov, V.(2017) Proc Natl Acad Sci U S A 114: 8211-8216
- PubMed: 28652344
- DOI: https://doi.org/10.1073/pnas.1702741114
- Primary Citation of Related Structures:
5VO8, 5VOI - PubMed Abstract:
Reiterative transcription is a noncanonical form of RNA synthesis in which a nucleotide specified by a single base in the DNA template is repetitively added to the nascent transcript. Here we determined the crystal structure of an RNA polymerase, the bacterial enzyme from Thermus thermophilus , engaged in reiterative transcription during transcription initiation at a promoter resembling the pyrG promoter of Bacillus subtilis The structure reveals that the reiterative transcript detours from the dedicated RNA exit channel and extends toward the main channel of the enzyme, thereby allowing RNA extension without displacement of the promoter recognition σ-factor. Nascent transcripts containing reiteratively added G residues are eventually extended by nonreiterative transcription, revealing an atypical pathway for the formation of a transcription elongation complex.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Center for RNA Molecular Biology, Pennsylvania State University, University Park, PA 16802; kum14@psu.edu vum5@psu.edu.