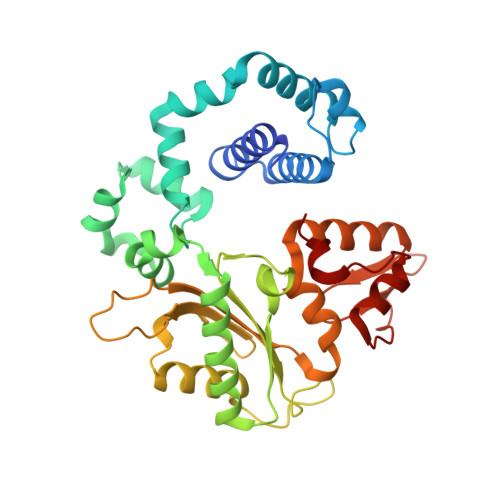
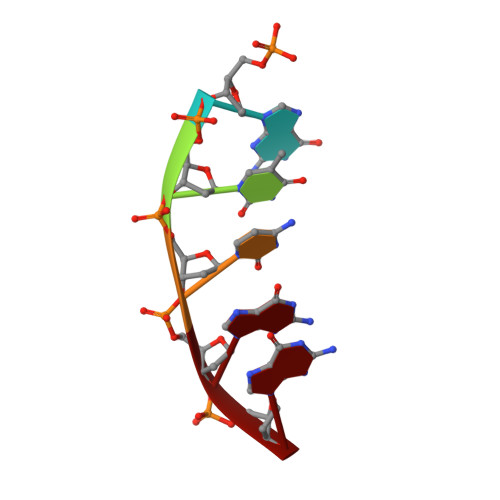
Capturing a mammalian DNA polymerase extending from an oxidized nucleotide.
Whitaker, A.M., Smith, M.R., Schaich, M.A., Freudenthal, B.D.(2017) Nucleic Acids Res 45: 6934-6944
- PubMed: 28449123
- DOI: https://doi.org/10.1093/nar/gkx293
- Primary Citation of Related Structures:
5V1F, 5V1G, 5V1H, 5V1I, 5V1J, 5V1N, 5V1O, 5V1P, 5V1R, 5VEZ - PubMed Abstract:
The oxidized nucleotide, 8-oxo-7,8-dihydro-2΄-deoxyguanosine (8-oxoG), is one of the most abundant DNA lesions. 8-oxoG plays a major role in tumorigenesis and human disease. Biological consequences of 8-oxoG are mediated in part by its insertion into the genome, making it essential to understand how DNA polymerases handle 8-oxoG. Insertion of 8-oxoG is mutagenic when opposite adenine but not when opposite cytosine. However, either result leads to DNA damage at the primer terminus (3΄-end) during the succeeding insertion event. Extension from DNA damage at primer termini remains poorly understood. Using kinetics and time-lapse crystallography, we evaluated how a model DNA polymerase, human polymerase β, accommodates 8-oxoG at the primer terminus opposite cytosine and adenine. Notably, extension from the mutagenic base pair is favored over the non-mutagenic base pair. When 8-oxoG is at the primer terminus opposite cytosine, DNA centric changes lead to a clash between O8 of 8-oxoG and the phosphate backbone. Changes in the extension reaction resulting from the altered active site provide evidence for a stabilizing interaction between Arg254 and Asp256 that serves an important role during DNA synthesis reactions. These results provide novel insights into the impact of damage at the primer terminus on genomic stability and DNA synthesis.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of Kansas Medical Center, Kansas City, KS 66160, USA.