Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Xu, J., Camacho, M., Xu, Y., Esser, V., Liu, X., Trimbuch, T., Pan, Y.Z., Ma, C., Tomchick, D.R., Rosenmund, C., Rizo, J.(2017) Elife 6
- PubMed: 28177287
- DOI: https://doi.org/10.7554/eLife.22567
- Primary Citation of Related Structures:
5UE8, 5UF7 - PubMed Abstract:
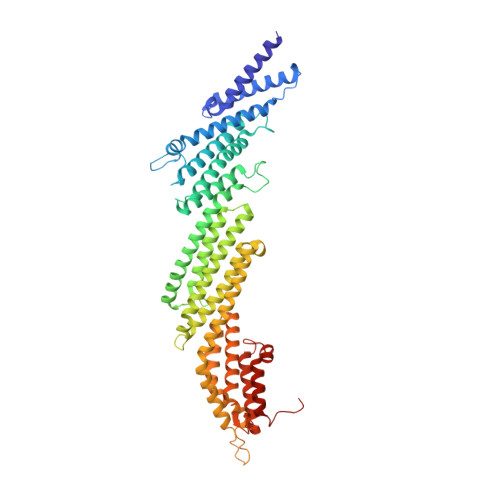
Munc13-1 acts as a master regulator of neurotransmitter release, mediating docking-priming of synaptic vesicles and diverse presynaptic plasticity processes. It is unclear how the functions of the multiple domains of Munc13-1 are coordinated. The crystal structure of a Munc13-1 fragment including its C 1 , C 2 B and MUN domains (C 1 C 2 BMUN) reveals a 19.5 nm-long multi-helical structure with the C 1 and C 2 B domains packed at one end. The similar orientations of the respective diacyglycerol- and Ca 2+ -binding sites of the C 1 and C 2 B domains suggest that the two domains cooperate in plasma-membrane binding and that activation of Munc13-1 by Ca 2+ and diacylglycerol during short-term presynaptic plasticity are closely interrelated. Electrophysiological experiments in mouse neurons support the functional importance of the domain interfaces observed in C 1 C 2 BMUN. The structure imposes key constraints for models of neurotransmitter release and suggests that Munc13-1 bridges the vesicle and plasma membranes from the periphery of the membrane-membrane interface.
Organizational Affiliation:
Department of Biophysics, University of Texas Southwestern Medical Center, Dallas, United States.