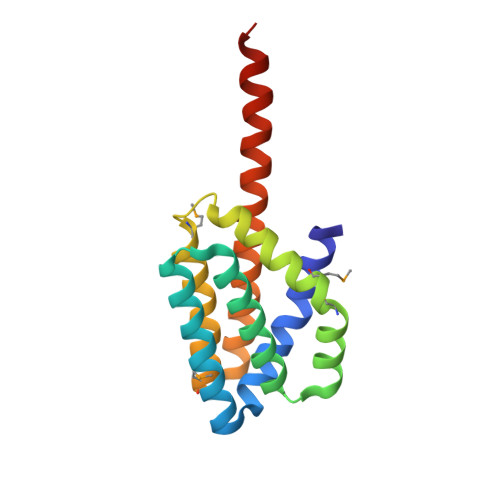
2.8 Angstrom Crystal Structure of the C-terminal Dimerization Domain of Transcriptional Regulator PdhR from Escherichia coli.
Minasov, G., Wawrzak, Z., Sandoval, J., Skarina, T., Grimshaw, S., Kwon, K., Savchenko, A., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.