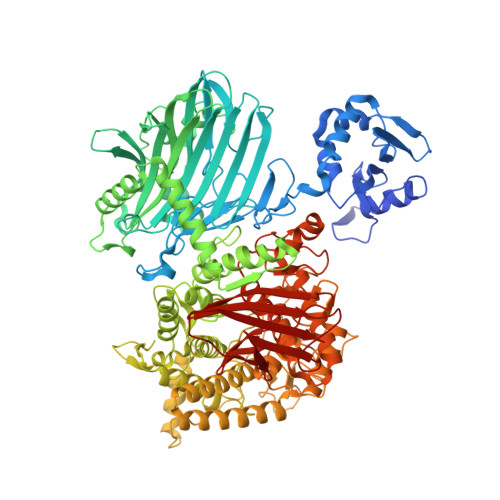
Cellodextrin phosphorylase from Ruminiclostridium thermocellum: X-ray crystal structure and substrate specificity analysis.
O'Neill, E.C., Pergolizzi, G., Stevenson, C.E.M., Lawson, D.M., Nepogodiev, S.A., Field, R.A.(2017) Carbohydr Res 451: 118-132
- PubMed: 28760417
- DOI: https://doi.org/10.1016/j.carres.2017.07.005
- Primary Citation of Related Structures:
5NZ7, 5NZ8 - PubMed Abstract:
The GH94 glycoside hydrolase cellodextrin phosphorylase (CDP, EC 2.4.1.49) produces cellodextrin oligomers from short β-1→4-glucans and α-D-glucose 1-phosphate. Compared to cellobiose phosphorylase (CBP), which produces cellobiose from glucose and α-D-glucose 1-phosphate, CDP is biochemically less well characterised. Herein, we investigate the donor and acceptor substrate specificity of recombinant CDP from Ruminiclostridium thermocellum and we isolate and characterise a glucosamine addition product to the cellobiose acceptor with the non-natural donor α-D-glucosamine 1-phosphate. In addition, we report the first X-ray crystal structure of CDP, along with comparison to the available structures from CBPs and other closely related enzymes, which contributes to understanding of the key structural features necessary to discriminate between monosaccharide (CBP) and oligosaccharide (CDP) acceptor substrates.
Organizational Affiliation:
Department of Biological Chemistry, John Innes Centre, Norwich Research Park, Norwich NR4 7UH, UK.