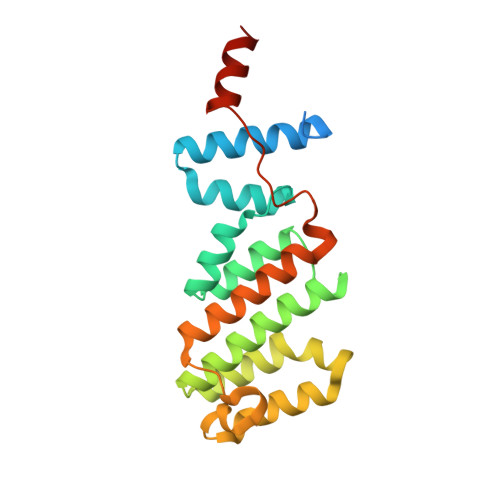
Structure of the Arabidopsis TOPLESS corepressor provides insight into the evolution of transcriptional repression.
Martin-Arevalillo, R., Nanao, M.H., Larrieu, A., Vinos-Poyo, T., Mast, D., Galvan-Ampudia, C., Brunoud, G., Vernoux, T., Dumas, R., Parcy, F.(2017) Proc Natl Acad Sci U S A 114: 8107-8112
- PubMed: 28698367
- DOI: https://doi.org/10.1073/pnas.1703054114
- Primary Citation of Related Structures:
5NQS, 5NQV - PubMed Abstract:
Transcriptional repression involves a class of proteins called corepressors that link transcription factors to chromatin remodeling complexes. In plants such as Arabidopsis thaliana , the most prominent corepressor is TOPLESS (TPL), which plays a key role in hormone signaling and development. Here we present the crystallographic structure of the Arabidopsis TPL N-terminal region comprising the LisH and CTLH (C-terminal to LisH) domains and a newly identified third region, which corresponds to a CRA domain. Comparing the structure of TPL with the mammalian TBL1, which shares a similar domain structure and performs a parallel corepressor function, revealed that the plant TPLs have evolved a new tetramerization interface and unique and highly conserved surface for interaction with repressors. Using site-directed mutagenesis, we validated those surfaces in vitro and in vivo and showed that TPL tetramerization and repressor binding are interdependent. Our results illustrate how evolution used a common set of protein domains to create a diversity of corepressors, achieving similar properties with different molecular solutions.
Organizational Affiliation:
Laboratoire de Physiologie Cellulaire et Végétale, Université Grenoble Alpes, CNRS, Commissariat à l'Energie Atomique et aux Energies Alternatives/Biosciences and Biotechnology Institute of Grenoble, Institut National de la Recherche Agronomique (INRA), F-38000 Grenoble, France.