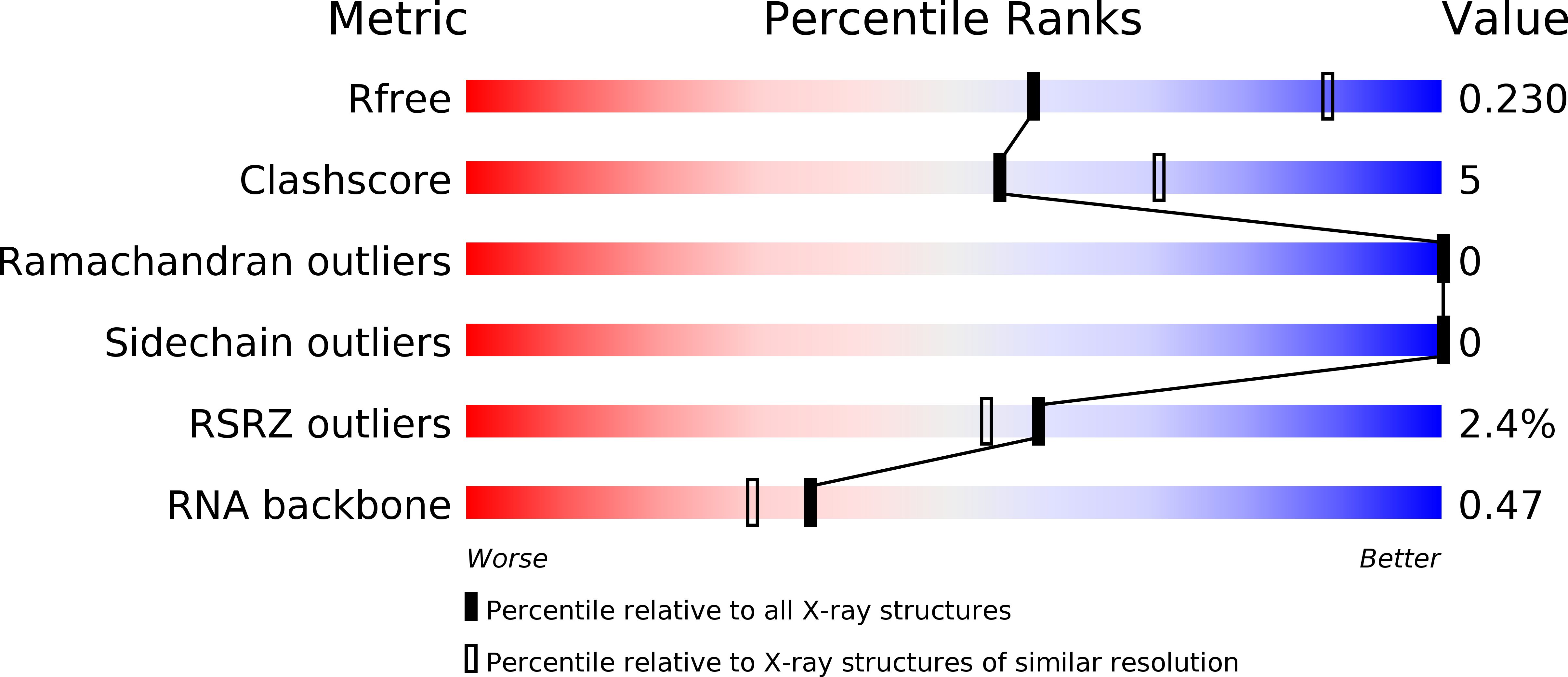
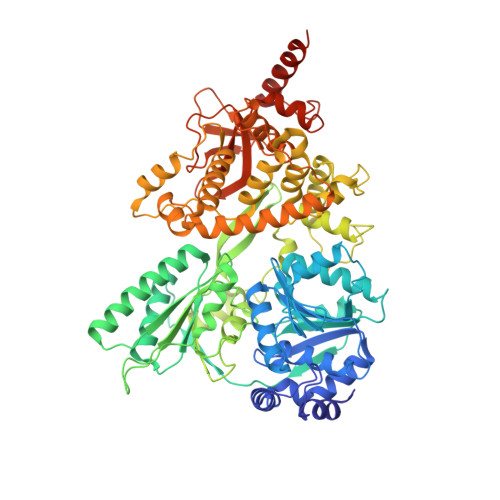
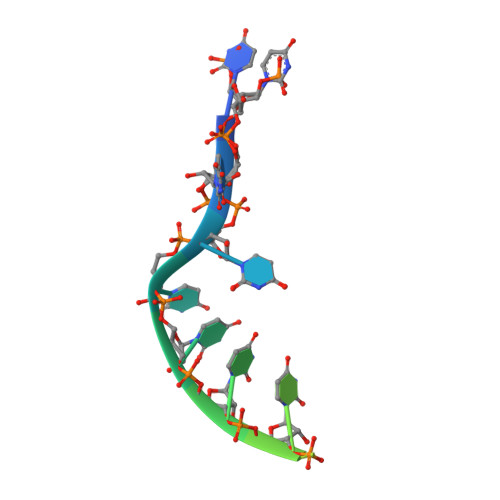
Structural insights into the mechanism of the DEAH-box RNA helicase Prp43.
Tauchert, M.J., Fourmann, J.B., Luhrmann, R., Ficner, R.(2017) Elife 6
- PubMed: 28092261
- DOI: https://doi.org/10.7554/eLife.21510
- Primary Citation of Related Structures:
5LTA, 5LTJ, 5LTK - PubMed Abstract:
The DEAH-box helicase Prp43 is a key player in pre-mRNA splicing as well as the maturation of rRNAs. The exact modus operandi of Prp43 and of all other spliceosomal DEAH-box RNA helicases is still elusive. Here, we report crystal structures of Prp43 complexes in different functional states and the analysis of structure-based mutants providing insights into the unwinding and loading mechanism of RNAs. The Prp43•ATP-analog•RNA complex shows the localization of the RNA inside a tunnel formed by the two RecA-like and C-terminal domains. In the ATP-bound state this tunnel can be transformed into a groove prone for RNA binding by large rearrangements of the C-terminal domains. Several conformational changes between the ATP- and ADP-bound states explain the coupling of ATP hydrolysis to RNA translocation, mainly mediated by a β-turn of the RecA1 domain containing the newly identified RF motif. This mechanism is clearly different to those of other RNA helicases.
Organizational Affiliation:
Department of Molecular Structural Biology, Institute for Microbiology and Genetics, GZMB, Georg-August-University Göttingen, Göttingen, Germany.