Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex.
Jiang, K., Rezabkova, L., Hua, S., Liu, Q., Capitani, G., Maarten Altelaar, A.F., Heck, A.J.R., Kammerer, R.A., Steinmetz, M.O., Akhmanova, A.(2017) Nat Cell Biol 19: 480-492
- PubMed: 28436967
- DOI: https://doi.org/10.1038/ncb3511
- Primary Citation of Related Structures:
5LB7, 5NBT - PubMed Abstract:
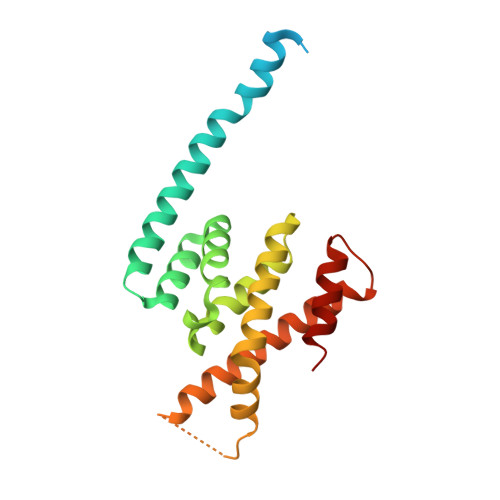
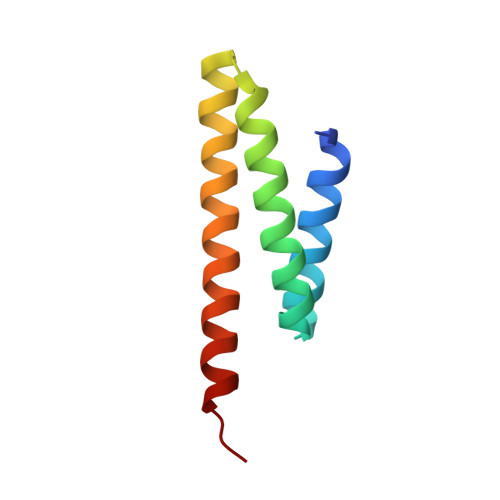
ASPM (known as Asp in fly and ASPM-1 in worm) is a microcephaly-associated protein family that regulates spindle architecture, but the underlying mechanism is poorly understood. Here, we show that ASPM forms a complex with another protein linked to microcephaly, the microtubule-severing ATPase katanin. ASPM and katanin localize to spindle poles in a mutually dependent manner and regulate spindle flux. X-ray crystallography revealed that the heterodimer formed by the N- and C-terminal domains of the katanin subunits p60 and p80, respectively, binds conserved motifs in ASPM. Reconstitution experiments demonstrated that ASPM autonomously tracks growing microtubule minus ends and inhibits their growth, while katanin decorates and bends both ends of dynamic microtubules and potentiates the minus-end blocking activity of ASPM. ASPM also binds along microtubules, recruits katanin and promotes katanin-mediated severing of dynamic microtubules. We propose that the ASPM-katanin complex controls microtubule disassembly at spindle poles and that misregulation of this process can lead to microcephaly.
Organizational Affiliation:
Cell Biology, Department of Biology, Faculty of Science, Utrecht University, Padualaan 8, 3584 CH Utrecht, The Netherlands.