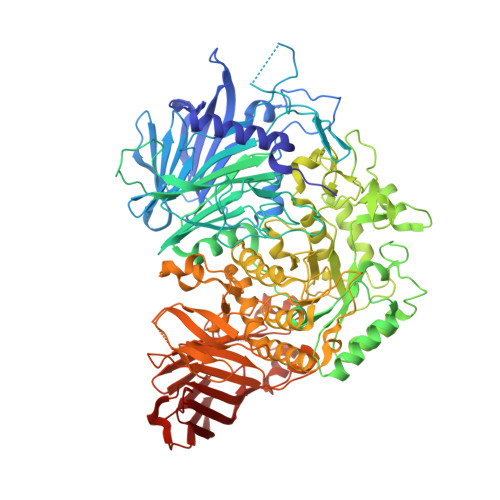
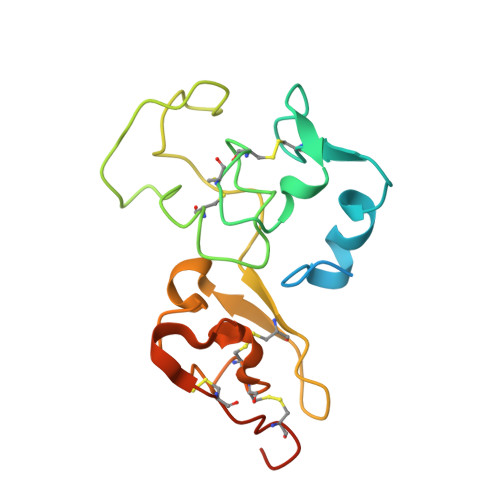
Interaction mode between catalytic and regulatory subunits in glucosidase II involved in ER glycoprotein quality control.
Satoh, T., Toshimori, T., Noda, M., Uchiyama, S., Kato, K.(2016) Protein Sci 25: 2095-2101
- PubMed: 27576940
- DOI: https://doi.org/10.1002/pro.3031
- Primary Citation of Related Structures:
5JQP - PubMed Abstract:
The glycoside hydrolase family 31 (GH31) α-glucosidases play vital roles in catabolic and regulated degradation, including the α-subunit of glucosidase II (GIIα), which catalyzes trimming of the terminal glucose residues of N-glycan in glycoprotein processing coupled with quality control in the endoplasmic reticulum (ER). Among the known GH31 enzymes, only GIIα functions with its binding partner, regulatory β-subunit (GIIβ), which harbors a lectin domain for substrate recognition. Although the structural data have been reported for GIIα and the GIIβ lectin domain, the interaction mode between GIIα and GIIβ remains unknown. Here, we determined the structure of a complex formed between GIIα and the GIIα-binding domain of GIIβ, thereby providing a structural basis underlying the functional extension of this unique GH31 enzyme.
Organizational Affiliation:
Graduate School of Pharmaceutical Sciences, Nagoya City University, 3-1 Tanabe-dori, Mizuho-ku, Nagoya, 467-8603, Japan. tadashisatoh@phar.nagoya-cu.ac.jp.