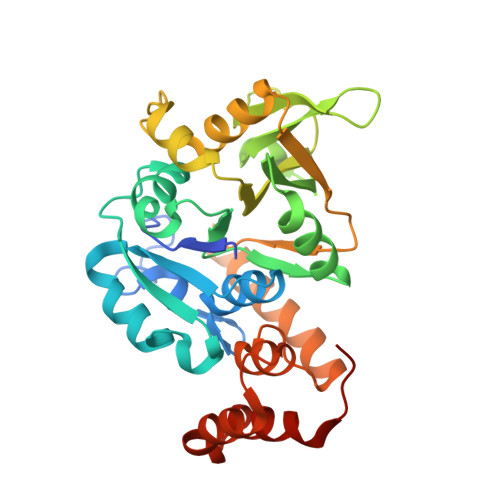
Crystal structure of Glucose-1-phosphate thymidylyltransferase from Burkholderia vietnamiensis in complex with 2'-Deoxyuridine-5'-monophosphate and 2'-Deoxy-Thymidine-B-L-Rhamnose
Abendroth, J., Dranow, D.M., Lorimer, D., Edwards, T.E.To be published.