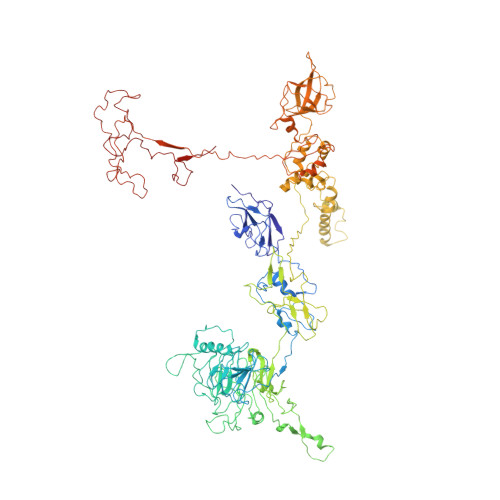
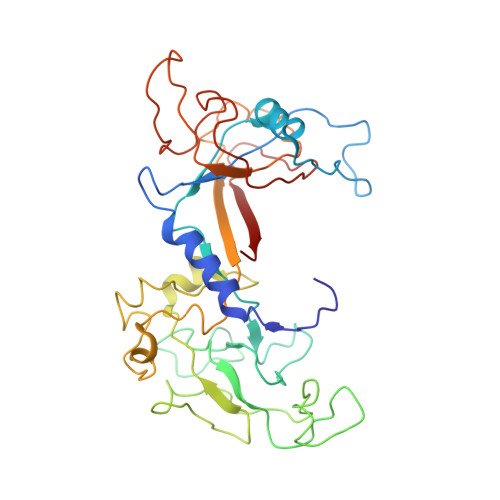
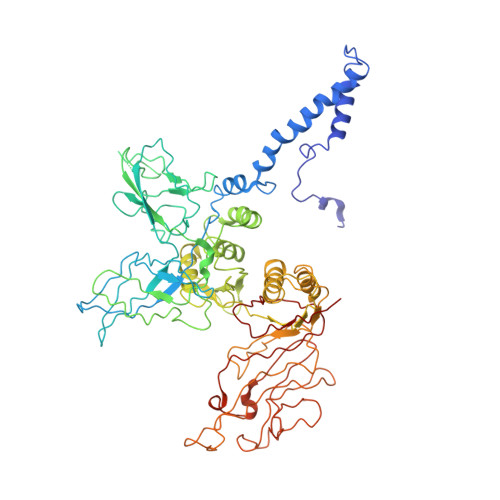
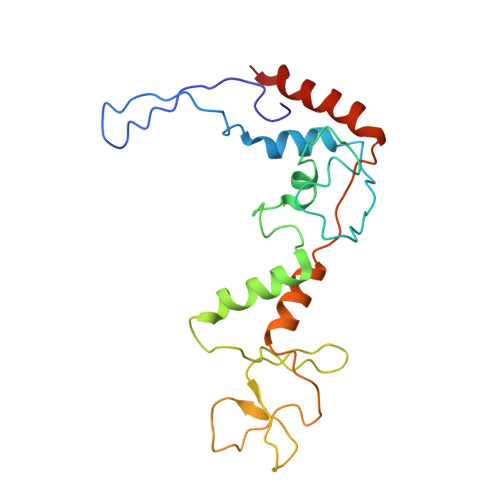
Role of bacteriophage T4 baseplate in regulating assembly and infection.
Yap, M.L., Klose, T., Arisaka, F., Speir, J.A., Veesler, D., Fokine, A., Rossmann, M.G.(2016) Proc Natl Acad Sci U S A 113: 2654-2659
- PubMed: 26929357
- DOI: https://doi.org/10.1073/pnas.1601654113
- Primary Citation of Related Structures:
5HX2 - PubMed Abstract:
Bacteriophage T4 consists of a head for protecting its genome and a sheathed tail for inserting its genome into a host. The tail terminates with a multiprotein baseplate that changes its conformation from a "high-energy" dome-shaped to a "low-energy" star-shaped structure during infection. Although these two structures represent different minima in the total energy landscape of the baseplate assembly, as the dome-shaped structure readily changes to the star-shaped structure when the virus infects a host bacterium, the dome-shaped structure must have more energy than the star-shaped structure. Here we describe the electron microscopy structure of a 3.3-MDa in vitro-assembled star-shaped baseplate with a resolution of 3.8 Å. This structure, together with other genetic and structural data, shows why the high-energy baseplate is formed in the presence of the central hub and how the baseplate changes to the low-energy structure, via two steps during infection. Thus, the presence of the central hub is required to initiate the assembly of metastable, high-energy structures. If the high-energy structure is formed and stabilized faster than the low-energy structure, there will be insufficient components to assemble the low-energy structure.
Organizational Affiliation:
Department of Biological Sciences, Purdue University, West Lafayette, IN 47907;