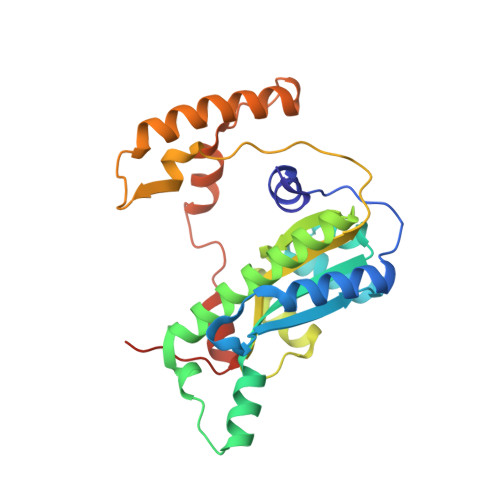
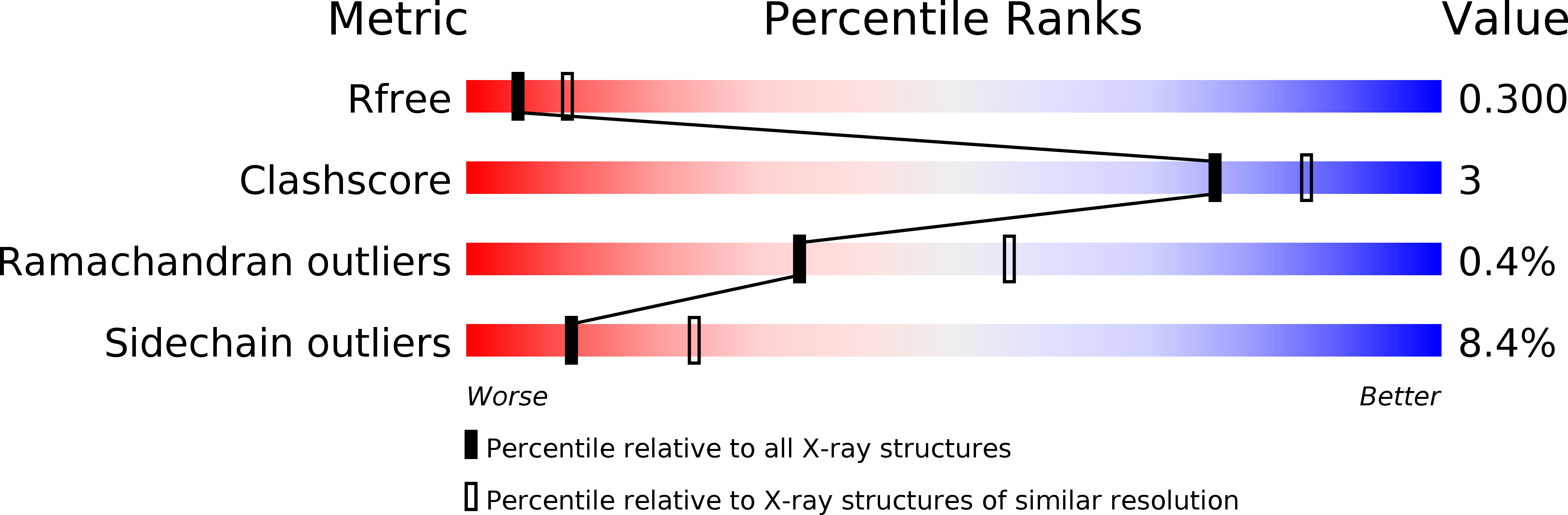Functional and structural characterization of two Bacillus megaterium nitroreductases biotransforming the herbicide mesotrione.
Carles, L., Besse-Hoggan, P., Joly, M., Vigouroux, A., Morera, S., Batisson, I.(2016) Biochem J 473: 1443-1453
- PubMed: 27005432
- DOI: https://doi.org/10.1042/BJ20151366
- Primary Citation of Related Structures:
5HDJ, 5HEI - PubMed Abstract:
Mesotrione is a selective herbicide belonging to the triketone family, commonly used on maize cultures since 2003. A mesotrione-transforming Bacillus megaterium Mes11 strain isolated from an agricultural soil was used as a model to identify the key enzymes initiating the biotransformation of this herbicide. Two enzymes (called NfrA1 and NfrA2/YcnD) were identified, and functionally and structurally characterized. Both belong to the NfsA FRP family of the nitro-FMN reductase superfamily (type I oxygen-insensitive nitroreductase) and show optimal pH and temperature of 6-6.5 and 23-25°C, respectively. Both undergo a Ping Pong Bi Bi mechanism, with NADPH and NADPH/NADH as cofactors for NfrA1 and NfrA2/YcnD, respectively. It is interesting that both can also reduce various nitro compounds including pesticides, antibiotics, one prodrug and 4-methylsulfonyl-2-nitrobenzoic acid, one of the mesotrione metabolites retrieved from the environment. The present study constitutes the first identification of mesotrione-transforming enzymes. These enzymes (or their corresponding genes) could be used as biomarkers to predict the capacity of ecosystems to transform mesotrione and assess their contamination by both the parent molecule and/or the metabolites.
Organizational Affiliation:
Clermont Université, Université Blaise Pascal, Laboratoire Microorganismes: Génome et Environnement (LMGE), F-63000 Clermont-Ferrand, France Centre National de la Recherche Scientifique (CNRS), UMR 6023, LMGE, TSA 60026, CS 60026, 63178 Aubière Cedex, France.