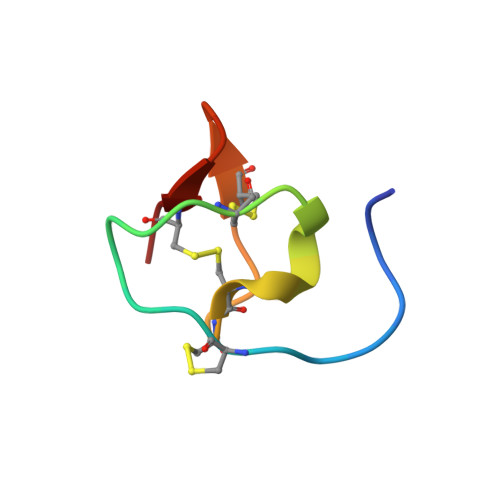
Characterization of Three Venom Peptides from the Spitting Spider Scytodes Thoracica.
Ariki, N.K., Munoz, L.E., Armitage, E.L., Goodstein, F.R., George, K.G., Smith, V.L., Vetter, I., Herzig, V., King, G.F., Loening, N.M.(2016) PLoS One 11: 56291
- PubMed: 27227898
- DOI: https://doi.org/10.1371/journal.pone.0156291
- Primary Citation of Related Structures:
5FZV, 5FZW, 5FZX - PubMed Abstract:
We present the solution-state NMR structures and preliminary functional characterizations of three venom peptides identified from the spitting spider Scytodes thoracica. Despite little sequence identity to other venom peptides, structural characterization reveals that these peptides contain an inhibitor cystine knot motif common to many venom peptides. These are the first structures for any peptide or protein from spiders of the Scytodidae family. Many venom peptides target neuronal ion channels or receptors. However, we have not been able to determine the target of these Scytodes peptides so we can only state with certainty the channels and receptors that they do not target.
Organizational Affiliation:
Chemistry Department, Lewis & Clark College, 0615 SW Palatine Hill Road, Portland, OR, 97219, United States of America.